Dynamic Crowding Regulates Transcription
- PMID: 31818468
- PMCID: PMC7202930
- DOI: 10.1016/j.bpj.2019.11.007
Dynamic Crowding Regulates Transcription
Abstract
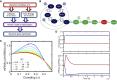
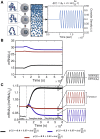
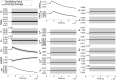
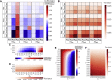
The nuclear environment is highly crowded by biological macromolecules, including chromatin and mobile proteins, which alter the kinetics and efficiency of transcriptional machinery. These alterations have been described, both theoretically and experimentally, for steady-state crowding densities; however, temporal changes in crowding density ("dynamic crowding") have yet to be integrated with gene expression. Dynamic crowding is pertinent to nuclear biology because processes such as chromatin translocation and protein diffusion lend to highly mobile biological crowders. Therefore, to capture such dynamic crowding and investigate its influence on transcription, we employ a three-pronged, systems-molecular approach. A system of chemical reactions represents the transcription pathway, the rates of which are determined by molecular-scale simulations; Brownian dynamics and Monte Carlo simulations quantify protein diffusion and DNA-protein binding affinity, dependent on macromolecular density. Altogether, this approach shows that transcription depends critically on dynamic crowding as the gene expression resultant from dynamic crowding can be profoundly different than that of steady-state crowding. In fact, expression levels can display both amplification and suppression and are notably different for genes or gene populations with different chemical and structural properties. These properties can be exploited to impose circadian expression, which is asymmetric and varies in strength, or to explain expression in cells under biomechanical stress. Therefore, this work demonstrates that dynamic crowding nontrivially alters transcription kinetics and presents dynamic crowding within the bulk nuclear nanoenvironment as a novel regulatory framework for gene expression.
Copyright © 2019 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Buldyrev S.V., Goldberger A.L., Stanley H.E. Long-range correlation properties of coding and noncoding DNA sequences: GenBank analysis. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1995;51:5084–5091. - PubMed
-
- Richter K., Nessling M., Lichter P. Experimental evidence for the influence of molecular crowding on nuclear architecture. J. Cell Sci. 2007;120:1673–1680. - PubMed
-
- Richter K., Nessling M., Lichter P. Macromolecular crowding and its potential impact on nuclear function. Biochim. Biophys. Acta. 2008;1783:2100–2107. - PubMed
-
- Al-Habori M. Macromolecular crowding and its role as intracellular signalling of cell volume regulation. Int. J. Biochem. Cell Biol. 2001;33:844–864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

