Distinct Genetic Signatures of Cortical and Subcortical Regions Associated with Human Memory
- PMID: 31818829
- PMCID: PMC6917897
- DOI: 10.1523/ENEURO.0283-19.2019
Distinct Genetic Signatures of Cortical and Subcortical Regions Associated with Human Memory
Abstract
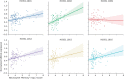
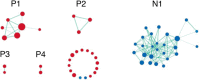
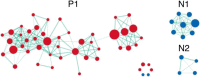
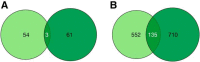
Despite the discovery of gene variants linked to memory performance, understanding the genetic basis of adult human memory remains a challenge. Here, we devised an unsupervised framework that relies on spatial correlations between human transcriptome data and functional neuroimaging maps to uncover the genetic signatures of memory in functionally-defined cortical and subcortical memory regions. Results were validated with animal literature and showed that our framework is highly effective in identifying memory-related processes and genes compared to a control cognitive function. Genes preferentially expressed in cortical memory regions are linked to memory-related processes such as immune and epigenetic regulation. Genes expressed in subcortical memory regions are associated with neurogenesis and glial cell differentiation. Genes expressed in both cortical and subcortical memory areas are involved in the regulation of transcription, synaptic plasticity, and glutamate receptor signaling. Furthermore, distinct memory-associated genes such as PRKCD and CDK5 are linked to cortical and subcortical regions, respectively. Thus, cortical and subcortical memory regions exhibit distinct genetic signatures that potentially reflect functional differences in health and disease, and nominates gene candidates for future experimental investigations.
Keywords: cognition; cortical; genetic; human; memory; neuroimaging.
Copyright © 2019 Tan et al.
Figures


Similar articles
-
Spatial analysis and high resolution mapping of the human whole-brain transcriptome for integrative analysis in neuroimaging.Neuroimage. 2018 Aug 1;176:259-267. doi: 10.1016/j.neuroimage.2018.04.068. Epub 2018 May 1. Neuroimage. 2018. PMID: 29723639
-
Evolutionary rate and gene expression across different brain regions.Genome Biol. 2008;9(9):R142. doi: 10.1186/gb-2008-9-9-r142. Genome Biol. 2008. PMID: 18811952 Free PMC article.
-
Beyond modularity: Fine-scale mechanisms and rules for brain network reconfiguration.Neuroimage. 2018 Feb 1;166:385-399. doi: 10.1016/j.neuroimage.2017.11.015. Epub 2017 Nov 11. Neuroimage. 2018. PMID: 29138087
-
The role of reelin in adult synaptic function and the genetic and epigenetic regulation of the reelin gene.Biochim Biophys Acta. 2008 Aug;1779(8):422-31. doi: 10.1016/j.bbagrm.2008.01.001. Epub 2008 Jan 12. Biochim Biophys Acta. 2008. PMID: 18237558 Review.
-
Neural bases of learning and memory: functional neuroimaging evidence.Curr Opin Neurol. 2000 Aug;13(4):415-21. doi: 10.1097/00019052-200008000-00008. Curr Opin Neurol. 2000. PMID: 10970058 Review.
Cited by
-
Decoding brain memory formation by single-cell RNA sequencing.Brief Bioinform. 2022 Nov 19;23(6):bbac412. doi: 10.1093/bib/bbac412. Brief Bioinform. 2022. PMID: 36156112 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
