MADS78 and MADS79 Are Essential Regulators of Early Seed Development in Rice
- PMID: 31818903
- PMCID: PMC6997703
- DOI: 10.1104/pp.19.00917
MADS78 and MADS79 Are Essential Regulators of Early Seed Development in Rice
Abstract
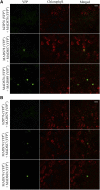
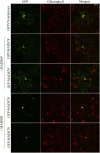
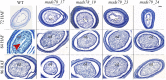
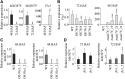
MADS box transcription factors (TFs) are subdivided into type I and II based on phylogenetic analysis. The type II TFs regulate floral organ identity and flowering time, but type I TFs are relatively less characterized. Here, we report the functional characterization of two type I MADS box TFs in rice (Oryza sativa), MADS78 and MADS79 Transcript abundance of both these genes in developing seed peaked at 48 h after fertilization and was suppressed by 96 h after fertilization, corresponding to syncytial and cellularized stages of endosperm development, respectively. Seeds overexpressing MADS78 and MADS 79 exhibited delayed endosperm cellularization, while CRISPR-Cas9-mediated single knockout mutants showed precocious endosperm cellularization. MADS78 and MADS 79 were indispensable for seed development, as a double knockout mutant failed to make viable seeds. Both MADS78 and 79 interacted with MADS89, another type I MADS box, which enhances nuclear localization. The expression analysis of Fie1, a rice FERTILIZATION-INDEPENDENT SEED-POLYCOMB REPRESSOR COMPLEX2 component, in MADS78 and 79 mutants and vice versa established an antithetical relation, suggesting that Fie1 could be involved in negative regulation of MADS78 and MADS 79 Misregulation of MADS78 and MADS 79 perturbed auxin homeostasis and carbon metabolism, as evident by misregulation of genes involved in auxin transport and signaling as well as starch biosynthesis genes causing structural abnormalities in starch granules at maturity. Collectively, we show that MADS78 and MADS 79 are essential regulators of early seed developmental transition and impact both seed size and quality in rice.
© 2020 American Society of Plant Biologists. All Rights Reserved.
Figures





Similar articles
-
Auxin production in the endosperm drives seed coat development in Arabidopsis.Elife. 2016 Nov 16;5:e20542. doi: 10.7554/eLife.20542. Elife. 2016. PMID: 27848912 Free PMC article.
-
The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1.Genes Dev. 2003 Jun 15;17(12):1540-53. doi: 10.1101/gad.257403. Genes Dev. 2003. PMID: 12815071 Free PMC article.
-
Rice fertilization-Independent Endosperm1 regulates seed size under heat stress by controlling early endosperm development.Plant Physiol. 2014 May;165(1):238-48. doi: 10.1104/pp.113.232413. Epub 2014 Mar 3. Plant Physiol. 2014. PMID: 24590858 Free PMC article.
-
The emerging importance of type I MADS box transcription factors for plant reproduction.Plant Cell. 2011 Mar;23(3):865-72. doi: 10.1105/tpc.110.081737. Epub 2011 Mar 4. Plant Cell. 2011. PMID: 21378131 Free PMC article. Review.
-
Function and diversification of MADS-box genes in rice.ScientificWorldJournal. 2006 Jul 6;6:1923-32. doi: 10.1100/tsw.2006.320. ScientificWorldJournal. 2006. PMID: 17205197 Free PMC article. Review.
Cited by
-
Allelic variation in rice Fertilization Independent Endosperm 1 contributes to grain width under high night temperature stress.New Phytol. 2021 Jan;229(1):335-350. doi: 10.1111/nph.16897. Epub 2020 Sep 23. New Phytol. 2021. PMID: 32858766 Free PMC article.
-
Temporal Control of Seed Development in Dicots: Molecular Bases, Ecological Impact and Possible Evolutionary Ramifications.Int J Mol Sci. 2021 Aug 26;22(17):9252. doi: 10.3390/ijms22179252. Int J Mol Sci. 2021. PMID: 34502157 Free PMC article. Review.
-
Natural variation in LONELY GUY-Like 1 regulates rice grain weight under warmer night conditions.Plant Physiol. 2024 Sep 2;196(1):164-180. doi: 10.1093/plphys/kiae313. Plant Physiol. 2024. PMID: 38820200 Free PMC article.
-
CTP synthase is essential for early endosperm development by regulating nuclei spacing.Plant Biotechnol J. 2021 Nov;19(11):2177-2191. doi: 10.1111/pbi.13644. Epub 2021 Jun 19. Plant Biotechnol J. 2021. PMID: 34058048 Free PMC article.
-
Bibliometric Analysis of Functional Crops and Nutritional Quality: Identification of Gene Resources to Improve Crop Nutritional Quality through Gene Editing Technology.Nutrients. 2023 Jan 11;15(2):373. doi: 10.3390/nu15020373. Nutrients. 2023. PMID: 36678244 Free PMC article.
References
-
- Abu-Zaitoon YM, Bennett K, Normanly J, Nonhebel HM (2012) A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiol Plant 146: 487–499 - PubMed
-
- Aguirre M, Kiegle E, Leo G, Ezquer I (2018) Carbohydrate reserves and seed development: An overview. Plant Reprod 31: 263–290 - PubMed
-
- Alexander MP. (1980) A versatile stain for pollen fungi, yeast and bacteria. Stain Technol 55: 13–18 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

