Pancreatic cancer organoids recapitulate disease and allow personalized drug screening
- PMID: 31818951
- PMCID: PMC6936689
- DOI: 10.1073/pnas.1911273116
Pancreatic cancer organoids recapitulate disease and allow personalized drug screening
Abstract
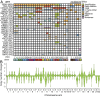
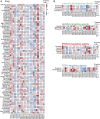
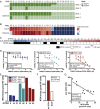
We report the derivation of 30 patient-derived organoid lines (PDOs) from tumors arising in the pancreas and distal bile duct. PDOs recapitulate tumor histology and contain genetic alterations typical of pancreatic cancer. In vitro testing of a panel of 76 therapeutic agents revealed sensitivities currently not exploited in the clinic, and underscores the importance of personalized approaches for effective cancer treatment. The PRMT5 inhibitor EZP015556, shown to target MTAP (a gene commonly lost in pancreatic cancer)-negative tumors, was validated as such, but also appeared to constitute an effective therapy for a subset of MTAP-positive tumors. Taken together, the work presented here provides a platform to identify novel therapeutics to target pancreatic tumor cells using PDOs.
Keywords: biobank; cancer; organoids; pancreatic cancer; personalized medicine.
Conflict of interest statement
Competing interest statement: H.C. is an inventor listed on several patents related to organoid technology. H.C. is the (unpaid) Chief Scientific Officer of Hubrecht Organoid Technology, a cofounder of Surrozen, and a scientific advisory board member for Kallyope, Merus, and Decibel. H. Clevers is a nonexecutive board member of Roche and Genentech and a scientific advisor for Life Sciences Partners. E.D. is an inventor on a patent related to organoid technology.
Figures


References
-
- Howlader N., Noone A., Krapcho M., Miller D., Bishop K., Kosary C. et al. , “SEER Cancer Statistics Review, 1975–2014” (National Cancer Institute, 2016).
-
- Conroy T., Desseigne F., Ychou M., Bouche O., Guimbaud R., Becouarn Y. et al. , FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl J Med. 364, 1817–1825 (2011). - PubMed
-
- Seino T., et al. , Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 22, 454–467.e6 (2018). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

