SIRT3 Haploinsufficiency Aggravates Loss of GABAergic Interneurons and Neuronal Network Hyperexcitability in an Alzheimer's Disease Model
- PMID: 31818974
- PMCID: PMC6961992
- DOI: 10.1523/JNEUROSCI.1446-19.2019
SIRT3 Haploinsufficiency Aggravates Loss of GABAergic Interneurons and Neuronal Network Hyperexcitability in an Alzheimer's Disease Model
Abstract
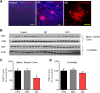
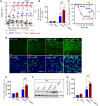
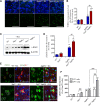
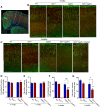
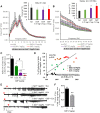
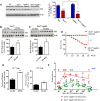
Impaired mitochondrial function and aberrant neuronal network activity are believed to be early events in the pathogenesis of Alzheimer's disease (AD), but how mitochondrial alterations contribute to aberrant activity in neuronal circuits is unknown. In this study, we examined the function of mitochondrial protein deacetylase sirtuin 3 (SIRT3) in the pathogenesis of AD. Compared with AppPs1 mice, Sirt3-haploinsufficient AppPs1 mice (Sirt3+/-AppPs1) exhibit early epileptiform EEG activity and seizure. Both male and female Sirt3+/-AppPs1 mice were observed to die prematurely before 5 months of age. When comparing male mice among different genotypes, Sirt3 haploinsufficiency renders GABAergic interneurons in the cerebral cortex vulnerable to degeneration and associated neuronal network hyperexcitability. Feeding Sirt3+/-AppPs1 AD mice with a ketone ester-rich diet increases SIRT3 expression and prevents seizure-related death and the degeneration of GABAergic neurons, indicating that the aggravated GABAergic neuron loss and neuronal network hyperexcitability in Sirt3+/-AppPs1 mice are caused by SIRT3 reduction and can be rescued by increase of SIRT3 expression. Consistent with a protective role in AD, SIRT3 levels are reduced in association with cerebral cortical Aβ pathology in AD patients. In summary, SIRT3 preserves GABAergic interneurons and protects cerebral circuits against hyperexcitability, and this neuroprotective mechanism can be bolstered by dietary ketone esters.SIGNIFICANCE STATEMENT GABAergic neurons provide the main inhibitory control of neuronal activity in the brain. By preserving mitochondrial function, SIRT3 protects parvalbumin and calretinin interneurons against Aβ-associated dysfunction and degeneration in AppPs1 Alzheimer's disease mice, thus restraining neuronal network hyperactivity. The neuronal network dysfunction that occurs in Alzheimer's disease can be partially reversed by physiological, dietary, and pharmacological interventions to increase SIRT3 expression and enhance the functionality of GABAergic interneurons.
Keywords: Alzheimer disease; GABAergic; hyperexcitability; mitochondria; seizure; telemetry.
Copyright © 2020 the authors.
Figures

Comment in
-
Seizing the Alzheimer's Brain: A Role for Sirtuin 3 in Hyperexcitability.Epilepsy Curr. 2020 Jun 30;20(4):224-226. doi: 10.1177/1535759720932157. eCollection 2020 Jul-Aug. Epilepsy Curr. 2020. PMID: 34025234 Free PMC article. No abstract available.
Similar articles
-
Involvement of GABAergic interneuron dysfunction and neuronal network hyperexcitability in Alzheimer's disease: Amelioration by metabolic switching.Int Rev Neurobiol. 2020;154:191-205. doi: 10.1016/bs.irn.2020.01.006. Epub 2020 Jul 10. Int Rev Neurobiol. 2020. PMID: 32739004 Review.
-
Mitochondrial SIRT3 Deficiency Results in Neuronal Network Hyperexcitability, Accelerates Age-Related Aβ Pathology, and Renders Neurons Vulnerable to Aβ Toxicity.Neuromolecular Med. 2023 Mar;25(1):27-39. doi: 10.1007/s12017-022-08713-2. Epub 2022 Jun 24. Neuromolecular Med. 2023. PMID: 35749057 Free PMC article.
-
Early alterations in hippocampal perisomatic GABAergic synapses and network oscillations in a mouse model of Alzheimer's disease amyloidosis.PLoS One. 2019 Jan 15;14(1):e0209228. doi: 10.1371/journal.pone.0209228. eCollection 2019. PLoS One. 2019. PMID: 30645585 Free PMC article.
-
SIRT3 mediates hippocampal synaptic adaptations to intermittent fasting and ameliorates deficits in APP mutant mice.Nat Commun. 2019 Apr 23;10(1):1886. doi: 10.1038/s41467-019-09897-1. Nat Commun. 2019. PMID: 31015456 Free PMC article.
-
Unsupervised excitation: GABAergic dysfunctions in Alzheimer's disease.Brain Res. 2019 Mar 15;1707:216-226. doi: 10.1016/j.brainres.2018.11.042. Epub 2018 Nov 29. Brain Res. 2019. PMID: 30503351 Review.
Cited by
-
GABAergic Inhibitory Interneuron Deficits in Alzheimer's Disease: Implications for Treatment.Front Neurosci. 2020 Jun 30;14:660. doi: 10.3389/fnins.2020.00660. eCollection 2020. Front Neurosci. 2020. PMID: 32714136 Free PMC article. Review.
-
Fats, Friends or Foes: Investigating the Role of Short- and Medium-Chain Fatty Acids in Alzheimer's Disease.Biomedicines. 2022 Nov 1;10(11):2778. doi: 10.3390/biomedicines10112778. Biomedicines. 2022. PMID: 36359298 Free PMC article. Review.
-
Parvalbumin Interneuron Dysfunction in Neurological Disorders: Focus on Epilepsy and Alzheimer's Disease.Int J Mol Sci. 2024 May 19;25(10):5549. doi: 10.3390/ijms25105549. Int J Mol Sci. 2024. PMID: 38791587 Free PMC article. Review.
-
Neuroregeneration Improved by Sodium-D,L-Beta-Hydroxybutyrate in Primary Neuronal Cultures.Pharmaceuticals (Basel). 2024 Aug 31;17(9):1160. doi: 10.3390/ph17091160. Pharmaceuticals (Basel). 2024. PMID: 39338322 Free PMC article.
-
Dementia in late-onset epilepsy: The Atherosclerosis Risk in Communities study.Neurology. 2020 Dec 15;95(24):e3248-e3256. doi: 10.1212/WNL.0000000000011080. Epub 2020 Oct 23. Neurology. 2020. PMID: 33097597 Free PMC article.
References
-
- Borchelt DR, Ratovitski T, van Lare J, Lee MK, Gonzales V, Jenkins NA, Copeland NG, Price DL, Sisodia SS (1997) Accelerated amyloid deposition in the brains of transgenic mice coexpressing mutant presenilin 1 and amyloid precursor proteins. Neuron 19:939–945. 10.1016/S0896-6273(00)80974-5 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
