Imaging Salt Uptake Dynamics in Plants Using PET
- PMID: 31819118
- PMCID: PMC6901586
- DOI: 10.1038/s41598-019-54781-z
Imaging Salt Uptake Dynamics in Plants Using PET
Abstract
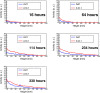
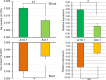
Soil salinity is a global environmental challenge for crop production. Understanding the uptake and transport properties of salt in plants is crucial to evaluate their potential for growth in high salinity soils and as a basis for engineering varieties with increased salt tolerance. Positron emission tomography (PET), traditionally used in medical and animal imaging applications for assessing and quantifying the dynamic bio-distribution of molecular species, has the potential to provide useful measurements of salt transport dynamics in an intact plant. Here we report on the feasibility of studying the dynamic transport of 22Na in millet using PET. Twenty-four green foxtail (Setaria viridis L. Beauv.) plants, 12 of each of two different accessions, were incubated in a growth solution containing 22Na+ ions and imaged at 5 time points over a 2-week period using a high-resolution small animal PET scanner. The reconstructed PET images showed clear evidence of sodium transport throughout the whole plant over time. Quantitative region-of-interest analysis of the PET data confirmed a strong correlation between total 22Na activity in the plants and time. Our results showed consistent salt transport dynamics within plants of the same variety and important differences between the accessions. These differences were corroborated by independent measurement of Na+ content and expression of the NHX transcript, a gene implicated in sodium transport. Our results demonstrate that PET can be used to quantitatively evaluate the transport of sodium in plants over time and, potentially, to discern differing salt-tolerance properties between plant varieties. In this paper, we also address the practical radiation safety aspects of working with 22Na in the context of plant imaging and describe a robust pipeline for handling and incubating plants. We conclude that PET is a promising and practical candidate technology to complement more traditional salt analysis methods and provide insights into systems-level salt transport mechanisms in intact plants.
Conflict of interest statement
The authors declare no competing interests.
Figures