A pair of E3 ubiquitin ligases compete to regulate filopodial dynamics and axon guidance
- PMID: 31820781
- PMCID: PMC7039193
- DOI: 10.1083/jcb.201902088
A pair of E3 ubiquitin ligases compete to regulate filopodial dynamics and axon guidance
Abstract
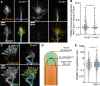
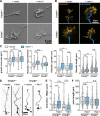
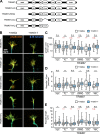
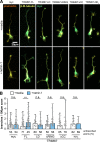
Appropriate axon guidance is necessary to form accurate neuronal connections. Axon guidance cues that stimulate cytoskeletal reorganization within the growth cone direct axon navigation. Filopodia at the growth cone periphery have long been considered sensors for axon guidance cues, yet how they respond to extracellular cues remains ill defined. Our previous work found that the filopodial actin polymerase VASP and consequently filopodial stability are negatively regulated via nondegradative TRIM9-dependent ubiquitination. Appropriate VASP ubiquitination and deubiquitination are required for axon turning in response to the guidance cue netrin-1. Here we show that the TRIM9-related protein TRIM67 outcompetes TRIM9 for interacting with VASP and antagonizes TRIM9-dependent VASP ubiquitination. The surprising antagonistic roles of two closely related E3 ubiquitin ligases are required for netrin-1-dependent filopodial responses, axon turning and branching, and fiber tract formation. We suggest a novel model in which coordinated regulation of VASP ubiquitination by a pair of interfering ligases is a critical element of VASP dynamics, filopodial stability, and axon guidance.
© 2019 Boyer et al.
Figures








References
-
- Bin J.M., Han D., Lai Wing Sun K., Croteau L.-P., Dumontier E., Cloutier J.-F., Kania A., and Kennedy T.E.. 2015. Complete Loss of Netrin-1 Results in Embryonic Lethality and Severe Axon Guidance Defects without Increased Neural Cell Death. Cell Reports. 12:1099–1106. 10.1016/j.celrep.2015.07.028 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

