Using Gene Editing to Establish a Safeguard System for Pluripotent Stem-Cell-Based Therapies
- PMID: 31821946
- PMCID: PMC6909005
- DOI: 10.1016/j.isci.2019.11.038
Using Gene Editing to Establish a Safeguard System for Pluripotent Stem-Cell-Based Therapies
Abstract
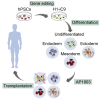
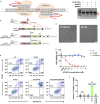
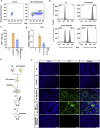
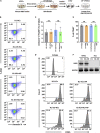
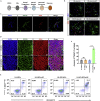
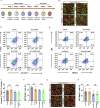
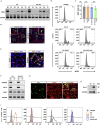
A major challenge in using human pluripotent stem cells (hPSCs) in therapy is the risk of teratoma formation due to contaminating undifferentiated stem cells. We used CRISPR-Cas9 for in-frame insertion of a suicide gene, iC9, into the endogenous SOX2 locus in human embryonic stem cell (ESC) line H1 for specific eradication of undifferentiated cells without affecting differentiated cells. This locus was chosen over NANOG and OCT4, two other well-characterized stem cell loci, due to significantly reduced off-target effect. We showed that undifferentiated H1-iC9 cells were induced to apoptosis by iC9 inducer AP1903, whereas differentiated cell lineages including hematopoietic cells, neurons, and islet beta-like cells were not affected. We also showed that AP1903 selectively removed undifferentiated H1-iC9 cells from a mixed cell population. This strategy therefore provides a layer of safety control before transplantation of a stem-cell-derived product in therapy.
Keywords: Cellular Therapy; Stem Cells Research; Techniques in Genetics.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors indicated no competing interests.
Figures
Similar articles
-
Inducible caspase-9 suicide gene under control of endogenous oct4 to safeguard mouse and human pluripotent stem cell therapy.Mol Ther Methods Clin Dev. 2022 Feb 1;24:332-341. doi: 10.1016/j.omtm.2022.01.014. eCollection 2022 Mar 10. Mol Ther Methods Clin Dev. 2022. PMID: 35229007 Free PMC article.
-
Sufficiency for inducible Caspase-9 safety switch in human pluripotent stem cells and disease cells.Gene Ther. 2019 Nov;27(10-11):525-534. doi: 10.1038/s41434-020-0179-z. Epub 2020 Jul 23. Gene Ther. 2019. PMID: 32704085
-
A Safeguard System for Induced Pluripotent Stem Cell-Derived Rejuvenated T Cell Therapy.Stem Cell Reports. 2015 Oct 13;5(4):597-608. doi: 10.1016/j.stemcr.2015.07.011. Epub 2015 Aug 28. Stem Cell Reports. 2015. PMID: 26321144 Free PMC article.
-
Human Induced Pluripotent Stem Cells in the Curative Treatment of Diabetes and Potential Impediments Ahead.Adv Exp Med Biol. 2019;1144:25-35. doi: 10.1007/5584_2018_305. Adv Exp Med Biol. 2019. PMID: 30569414 Review.
-
CRISPR/Cas9 genome editing in human pluripotent stem cells: Harnessing human genetics in a dish.Dev Dyn. 2016 Jul;245(7):788-806. doi: 10.1002/dvdy.24414. Epub 2016 Jun 9. Dev Dyn. 2016. PMID: 27145095 Review.
Cited by
-
Strategies to Improve the Safety of iPSC-Derived β Cells for β Cell Replacement in Diabetes.Transpl Int. 2022 Aug 24;35:10575. doi: 10.3389/ti.2022.10575. eCollection 2022. Transpl Int. 2022. PMID: 36090777 Free PMC article. Review.
-
Salicylic diamines selectively eliminate residual undifferentiated cells from pluripotent stem cell-derived cardiomyocyte preparations.Sci Rep. 2021 Jan 27;11(1):2391. doi: 10.1038/s41598-021-81351-z. Sci Rep. 2021. PMID: 33504837 Free PMC article.
-
Scalable manufacturing of clinical-grade differentiated cardiomyocytes derived from human-induced pluripotent stem cells for regenerative therapy.Cell Prolif. 2022 Aug;55(8):e13248. doi: 10.1111/cpr.13248. Epub 2022 May 9. Cell Prolif. 2022. PMID: 35534945 Free PMC article. Review.
-
The Use of MSCs, iPSCs, and EVs in the Repair of Human MSK Tissues: Is Ultimate Success Dependent on Developing Excellent Implant Materials as Well as Creating an Optimal Environment for Implantation? What Is the Rationale for These Choices?Int J Mol Sci. 2025 Jun 28;26(13):6250. doi: 10.3390/ijms26136250. Int J Mol Sci. 2025. PMID: 40650028 Free PMC article. Review.
-
Inducible caspase-9 suicide gene under control of endogenous oct4 to safeguard mouse and human pluripotent stem cell therapy.Mol Ther Methods Clin Dev. 2022 Feb 1;24:332-341. doi: 10.1016/j.omtm.2022.01.014. eCollection 2022 Mar 10. Mol Ther Methods Clin Dev. 2022. PMID: 35229007 Free PMC article.
References
-
- Balboa D., Weltner J., Novik Y., Eurola S., Wartiovaara K., Otonkoski T. Generation of an OCT4 reporter human induced pluripotent stem cell line using CRISPR/SpCas9. Stem Cell Res. 2017;23:105–108. - PubMed
-
- Bonini C., Ferrari G., Verzeletti S., Servida P., Zappone E., Ruggieri L., Ponzoni M., Rossini S., Mavilio F., Traversari C. HSV-TK gene transfer into donor lymphocytes for control of allogeneic graft-versus-leukemia. Science. 1997;276:1719–1724. - PubMed
-
- Caserta T.M., Smith A.N., Gultice A.D., Reedy M.A., Brown T.L. Q-VD-OPh, a broad spectrum caspase inhibitor with potent antiapoptotic properties. Apoptosis. 2003;8:345–352. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

