Investigating RNA expression profiles altered by nicotinamide mononucleotide therapy in a chronic model of alcoholic liver disease
- PMID: 31823815
- PMCID: PMC6902345
- DOI: 10.1186/s40246-019-0251-1
Investigating RNA expression profiles altered by nicotinamide mononucleotide therapy in a chronic model of alcoholic liver disease
Abstract
Background: Chronic alcohol consumption is a significant cause of liver disease worldwide. Several biochemical mechanisms have been linked to the initiation and progression of alcoholic liver disease (ALD) such as oxidative stress, inflammation, and metabolic dysregulation, including the disruption of NAD+/NADH. Indeed, an ethanol-mediated reduction in hepatic NAD+ levels is thought to be one factor underlying ethanol-induced steatosis, oxidative stress, steatohepatitis, insulin resistance, and inhibition of gluconeogenesis. Therefore, we applied a NAD+ boosting supplement to investigate alterations in the pathogenesis of early-stage ALD.
Methods: To examine the impact of NAD+ therapy on the early stages of ALD, we utilized nicotinamide mononucleotide (NMN) at 500 mg/kg intraperitoneal injection every other day, for the duration of a Lieber-DeCarli 6-week chronic ethanol model in mice. Numerous strategies were employed to characterize the effect of NMN therapy, including the integration of RNA-seq, immunoblotting, and metabolomics analysis.
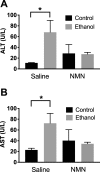
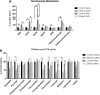
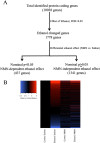
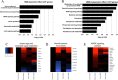
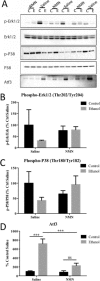
Results: Our findings reveal that NMN therapy increased hepatic NAD+ levels, prevented an ethanol-induced increase in plasma ALT and AST, and changed the expression of 25% of the genes that were modulated by ethanol metabolism. These genes were associated with a number of pathways including the MAPK pathway. Interestingly, our analysis revealed that NMN treatment normalized Erk1/2 signaling and prevented an induction of Atf3 overexpression.
Conclusions: These findings reveal previously unreported mechanisms by which NMN supplementation alters hepatic gene expression and protein pathways to impact ethanol hepatotoxicity in an early-stage murine model of ALD. Overall, our data suggest further research is needed to fully characterize treatment paradigms and biochemical implications of NAD+-based interventions.
Keywords: ATF3; Alcoholic liver disease; ERK1/2; Liver; NMN; RNA-seq; Sirtuin.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

