Comparison Study of Computational Prediction Tools for Drug-Target Binding Affinities
- PMID: 31824921
- PMCID: PMC6879652
- DOI: 10.3389/fchem.2019.00782
Comparison Study of Computational Prediction Tools for Drug-Target Binding Affinities
Abstract
The drug development is generally arduous, costly, and success rates are low. Thus, the identification of drug-target interactions (DTIs) has become a crucial step in early stages of drug discovery. Consequently, developing computational approaches capable of identifying potential DTIs with minimum error rate are increasingly being pursued. These computational approaches aim to narrow down the search space for novel DTIs and shed light on drug functioning context. Most methods developed to date use binary classification to predict if the interaction between a drug and its target exists or not. However, it is more informative but also more challenging to predict the strength of the binding between a drug and its target. If that strength is not sufficiently strong, such DTI may not be useful. Therefore, the methods developed to predict drug-target binding affinities (DTBA) are of great value. In this study, we provide a comprehensive overview of the existing methods that predict DTBA. We focus on the methods developed using artificial intelligence (AI), machine learning (ML), and deep learning (DL) approaches, as well as related benchmark datasets and databases. Furthermore, guidance and recommendations are provided that cover the gaps and directions of the upcoming work in this research area. To the best of our knowledge, this is the first comprehensive comparison analysis of tools focused on DTBA with reference to AI/ML/DL.
Keywords: artificial intelligence; bioinformatics; deep learning; drug repurposing; drug-target binding affinity; drug-target interaction; information integration; machine learning.
Copyright © 2019 Thafar, Raies, Albaradei, Essack and Bajic.
Figures
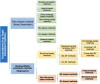

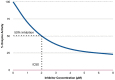
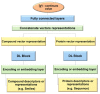
References
-
- Alshahrani M., Hoehndorf R. (2018). Drug repurposing through joint learning on knowledge graphs and literature. bioRXiv [Preprint]. 10.1101/385617 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

