Comprehensive Identification and Characterization of Human Secretome Based on Integrative Proteomic and Transcriptomic Data
- PMID: 31824949
- PMCID: PMC6881247
- DOI: 10.3389/fcell.2019.00299
Comprehensive Identification and Characterization of Human Secretome Based on Integrative Proteomic and Transcriptomic Data
Abstract
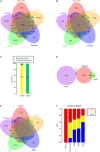
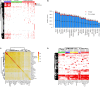
Secreted proteins (SPs) play important roles in diverse important biological processes; however, a comprehensive and high-quality list of human SPs is still lacking. Here we identified 6,943 high-confidence human SPs (3,522 of them are novel) based on 330,427 human proteins derived from databases of UniProt, Ensembl, AceView, and RefSeq. Notably, 6,267 of 6,943 (90.3%) SPs have the supporting evidences from a large amount of mass spectrometry (MS) and RNA-seq data. We found that the SPs were broadly expressed in diverse tissues as well as human body fluid, and a significant portion of them exhibited tissue-specific expression. Moreover, 14 cancer-specific SPs that their expression levels were significantly associated with the patients' survival of eight different tumors were identified, which could be potential prognostic biomarkers. Strikingly, 89.21% of 6,943 SPs (2,927 novel SPs) contain known protein domains. Those novel SPs we mainly enriched with the known domains regarding immunity, such as Immunoglobulin V-set and C1-set domain. Specifically, we constructed a user-friendly and freely accessible database, SPRomeDB (www.unimd.org/SPRomeDB), to catalog those SPs. Our comprehensive SP identification and characterization gain insights into human secretome and provide valuable resource for future researches.
Keywords: RNA-seq; human secretome; proteome; secreted proteins; transcriptome.
Copyright © 2019 Chen, Chen, Liu, Chen, Zhang, Li, Thierry-Mieg, Thierry-Mieg, Mattes, Ning and Shi.
Figures



References
LinkOut - more resources
Full Text Sources

