Optimized Combination of Multiple Graphs With Application to the Integration of Brain Imaging and (epi)Genomics Data
- PMID: 31825864
- PMCID: PMC7394342
- DOI: 10.1109/TMI.2019.2958256
Optimized Combination of Multiple Graphs With Application to the Integration of Brain Imaging and (epi)Genomics Data
Abstract
With the rapid development of high-throughput technologies, a growing amount of multi-omics data are collected, giving rise to a great demand for combining such data for biomedical discovery. Due to the cost and time to label the data manually, the number of labelled samples is limited. This motivated the need for semi-supervised learning algorithms. In this work, we applied a graph-based semi-supervised learning (GSSL) to classify a severe chronic mental disorder, schizophrenia (SZ). An advantage of GSSL is that it can simultaneously analyse more than two types of data, while many existing models focus on pairwise data analysis. In particular, we applied GSSL to the analysis of single nucleotide polymorphism (SNP), functional magnetic resonance imaging (fMRI) and DNA methylation data, which accounts for genetics, brain imaging (endophenotypes), and environmental factors (epigenomics) respectively. While parameter selection has been an open challenge for most models, another key contribution of this work is that we explored the parameter space to interpret their meaning and established practical guidelines. Based on the practical significance of each hyper-parameter, a relatively small range of candidate values can be determined in a data-driven way to both optimize and speed up the parameter tuning process. We validated the model through both synthetic data and a real SZ dataset of 184 subjects from the Mental Illness and Neuroscience Discovery (MIND) Clinical Imaging Consortium. In comparison to several existing approaches, our algorithm achieved better performance in terms of classification accuracy. We also confirmed the significance of several brain regions associated with SZ.
Figures

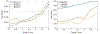

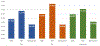



References
-
- Miao R, Luo H, Zhou H, Li G, Bu D, Yang X, et al. Identification of prognostic biomarkers in hepatitis b virus-related hepatocellular carcinoma and stratification by integrative multi-omics analysis. Journal of hepatology, 61(4):840–849, 2014. - PubMed
-
- Cisek K, Krochmal M, Klein J, and Mischak H The application of multi-omics and systems biology to identify therapeutic targets in chronic kidney disease. Nephrology Dialysis Transplantation, 31(12):2003–2011, 2015. - PubMed
-
- Wheelock CE, Goss VM, Balgoma D, Nicholas B, Brandsma J, Skipp PJ, et al. Application of omics technologies to biomarker discovery in inflammatory lung diseases. European Respiratory Journal, 42(3):802–825, 2013. - PubMed

