Clinical Implications of Sub-grouping HER2 Positive Tumors by Amplicon Structure and Co-amplified Genes
- PMID: 31827209
- PMCID: PMC6906288
- DOI: 10.1038/s41598-019-55455-6
Clinical Implications of Sub-grouping HER2 Positive Tumors by Amplicon Structure and Co-amplified Genes
Erratum in
-
Author Correction: Clinical Implications of Sub-grouping HER2 Positive Tumors by Amplicon Structure and Co-amplified Genes.Sci Rep. 2020 Feb 27;10(1):3941. doi: 10.1038/s41598-020-60492-7. Sci Rep. 2020. PMID: 32109238 Free PMC article.
Abstract
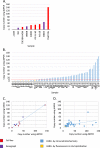
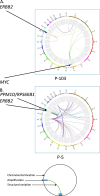
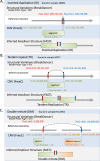
ERBB2 amplification is a prognostic marker for aggressive tumors and a predictive marker for prolonged survival following treatment with HER2 inhibitors. We attempt to sub-group HER2+ tumors based on amplicon structures and co-amplified genes. We examined five HER2+ cell lines, three HER2+ xenographs and 57 HER2+ tumor tissues. ERBB2 amplification was analyzed using digital droplet PCR and low coverage whole genome sequencing. In some HER2+ tumors PPM1D, that encodes WIP1, is co-amplified. Cell lines were treated with HER2 and WIP1 inhibitors. We find that inverted duplication is the amplicon structure in the majority of HER2+ tumors. In patients suffering from an early stage disease the ERBB2 amplicon is composed of a single segment while in patients suffering from advanced cancer the amplicon is composed of several different segments. We find robust WIP1 inhibition in some HER2+ PPM1D amplified cell lines. Sub-grouping HER2+ tumors using low coverage whole genome sequencing identifies inverted duplications as the main amplicon structure and based on the number of segments, differentiates between local and advanced tumors. In addition, we found that we could determine if a tumor is a recurrent tumor or second primary tumor and identify co-amplified oncogenes that may serve as targets for therapy.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bang YJ, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

