KRAS4A directly regulates hexokinase 1
- PMID: 31827279
- PMCID: PMC6923592
- DOI: 10.1038/s41586-019-1832-9
KRAS4A directly regulates hexokinase 1
Abstract
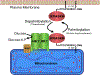
The most frequently mutated oncogene in cancer is KRAS, which uses alternative fourth exons to generate two gene products (KRAS4A and KRAS4B) that differ only in their C-terminal membrane-targeting region1. Because oncogenic mutations occur in exons 2 or 3, two constitutively active KRAS proteins-each capable of transforming cells-are encoded when KRAS is activated by mutation2. No functional distinctions among the splice variants have so far been established. Oncogenic KRAS alters the metabolism of tumour cells3 in several ways, including increased glucose uptake and glycolysis even in the presence of abundant oxygen4 (the Warburg effect). Whereas these metabolic effects of oncogenic KRAS have been explained by transcriptional upregulation of glucose transporters and glycolytic enzymes3-5, it is not known whether there is direct regulation of metabolic enzymes. Here we report a direct, GTP-dependent interaction between KRAS4A and hexokinase 1 (HK1) that alters the activity of the kinase, and thereby establish that HK1 is an effector of KRAS4A. This interaction is unique to KRAS4A because the palmitoylation-depalmitoylation cycle of this RAS isoform enables colocalization with HK1 on the outer mitochondrial membrane. The expression of KRAS4A in cancer may drive unique metabolic vulnerabilities that can be exploited therapeutically.
Conflict of interest statement
Figures

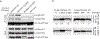
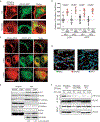


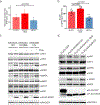

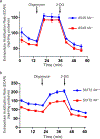





References
-
- Voice JK, Klemke RL, Le A & Jackson JH Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J Biol Chem 274, 17164–17170. (1999). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 CA009161/CA/NCI NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- R01 CA116034/CA/NCI NIH HHS/United States
- R35 CA232124/CA/NCI NIH HHS/United States
- R01 CA214948/CA/NCI NIH HHS/United States
- P01 CA117969/CA/NCI NIH HHS/United States
- R01 CA163489/CA/NCI NIH HHS/United States
- U01 CA217864/CA/NCI NIH HHS/United States
- R01 CA157490/CA/NCI NIH HHS/United States
- R01 GM095567/GM/NIGMS NIH HHS/United States
- R35 CA210018/CA/NCI NIH HHS/United States
- R01 CA188048/CA/NCI NIH HHS/United States
- P41 EB017183/EB/NIBIB NIH HHS/United States
- T32 GM088118/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

