Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease
- PMID: 31827281
- PMCID: PMC6930849
- DOI: 10.1038/s41586-019-1828-5
Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease
Abstract
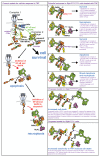
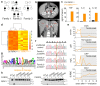
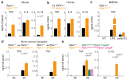
RIPK1 is a key regulator of innate immune signalling pathways. To ensure an optimal inflammatory response, RIPK1 is regulated post-translationally by well-characterized ubiquitylation and phosphorylation events, as well as by caspase-8-mediated cleavage1-7. The physiological relevance of this cleavage event remains unclear, although it is thought to inhibit activation of RIPK3 and necroptosis8. Here we show that the heterozygous missense mutations D324N, D324H and D324Y prevent caspase cleavage of RIPK1 in humans and result in an early-onset periodic fever syndrome and severe intermittent lymphadenopathy-a condition we term 'cleavage-resistant RIPK1-induced autoinflammatory syndrome'. To define the mechanism for this disease, we generated a cleavage-resistant Ripk1D325A mutant mouse strain. Whereas Ripk1-/- mice died postnatally from systemic inflammation, Ripk1D325A/D325A mice died during embryogenesis. Embryonic lethality was completely prevented by the combined loss of Casp8 and Ripk3, but not by loss of Ripk3 or Mlkl alone. Loss of RIPK1 kinase activity also prevented Ripk1D325A/D325A embryonic lethality, although the mice died before weaning from multi-organ inflammation in a RIPK3-dependent manner. Consistently, Ripk1D325A/D325A and Ripk1D325A/+ cells were hypersensitive to RIPK3-dependent TNF-induced apoptosis and necroptosis. Heterozygous Ripk1D325A/+ mice were viable and grossly normal, but were hyper-responsive to inflammatory stimuli in vivo. Our results demonstrate the importance of caspase-mediated RIPK1 cleavage during embryonic development and show that caspase cleavage of RIPK1 not only inhibits necroptosis but also maintains inflammatory homeostasis throughout life.
Conflict of interest statement
Figures








References
-
- Bertrand MJM, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. - PubMed
-
- Dondelinger Y, et al. MK2 phosphorylation of RIPK1 regulates TNF-mediated cell death. Nat Cell Biol. 2017;19:1237–1247. - PubMed
-
- Dondelinger Y, et al. NF-kappaB-Independent Role of IKKalpha/IKKbeta in Preventing RIPK1 Kinase-Dependent Apoptotic and Necroptotic Cell Death during TNF Signaling. Mol Cell. 2015;60:63–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

