NANUQ: a method for inferring species networks from gene trees under the coalescent model
- PMID: 31827592
- PMCID: PMC6896299
- DOI: 10.1186/s13015-019-0159-2
NANUQ: a method for inferring species networks from gene trees under the coalescent model
Abstract
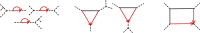
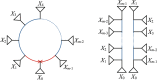
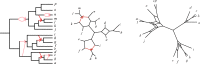
Species networks generalize the notion of species trees to allow for hybridization or other lateral gene transfer. Under the network multispecies coalescent model, individual gene trees arising from a network can have any topology, but arise with frequencies dependent on the network structure and numerical parameters. We propose a new algorithm for statistical inference of a level-1 species network under this model, from data consisting of gene tree topologies, and provide the theoretical justification for it. The algorithm is based on an analysis of quartets displayed on gene trees, combining several statistical hypothesis tests with combinatorial ideas such as a quartet-based intertaxon distance appropriate to networks, the NeighborNet algorithm for circular split systems, and the Circular Network algorithm for constructing a splits graph.
Keywords: Gene tree; Hybridization; Level-1 network; NANUQ; Network multispecies coalescent; Quartets; Species network inference.
© The Author(s) 2019.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures











References
Grants and funding
LinkOut - more resources
Full Text Sources

