A Genomic-Clinicopathologic Nomogram Predicts Survival for Patients with Laryngeal Squamous Cell Carcinoma
- PMID: 31827637
- PMCID: PMC6886334
- DOI: 10.1155/2019/5980567
A Genomic-Clinicopathologic Nomogram Predicts Survival for Patients with Laryngeal Squamous Cell Carcinoma
Abstract
Background: Long noncoding RNAs (lncRNAs), which have little or no ability to encode proteins, have attracted special attention due to their potential role in cancer disease. We aimed to establish a lncRNA signature and a nomogram incorporating the genomic and clinicopathologic factors to improve the accuracy of survival prediction for laryngeal squamous cell carcinoma (LSCC).
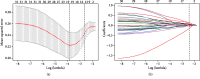
Methods: A LSCC RNA-sequencing (RNA-seq) dataset and the matched clinicopathologic information were downloaded from The Cancer Genome Atlas (TCGA). Using univariable Cox regression and least absolute shrinkage and selection operator (LASSO) analysis, we developed a thirteen-lncRNA signature related to prognosis. On the basis of multivariable Cox regression analysis results, a nomogram integrating the genomic and clinicopathologic predictors was built. The predictive accuracy and discriminative ability of the inclusive nomogram were confirmed by calibration curve and a concordance index (C-index), and compared with the TNM staging system by C-index and receiver operating characteristic (ROC) analysis. Decision curve analysis (DCA) was conducted to evaluate the clinical value of our nomogram.
Results: Thirteen overall survival- (OS-) related lncRNAs were identified, and the signature consisting of the selected thirteen lncRNAs could effectively divide patients into high-risk and low-risk subgroups, with area under curves (AUC) of 0.89 (3-year OS) and 0.885 (5-year OS). Independent factors derived from multivariable analysis to predict survival were margin status, tumor status, and lncRNA signature, which were all assembled into the nomogram. The calibration curve for the survival probability showed that the predictions based on the nomogram coincided well with actual observations. The C-index of the nomogram was 0.82 (0.77-0.87), and the area under curve (AUC) of the nomogram in predicting overall survival (OS) was 0.938, both of which were significantly higher than the traditional TNM stage. Decision curve analysis further demonstrated that our nomogram had larger net benefit than TNM stage.
Conclusion: An inclusive nomogram for patients with LSCC, comprising genomic and clinicopathologic variables, generates more accurate estimations of the survival probability when compared with TNM stage alone, but more data are needed before the nomogram is used in clinical practice.
Copyright © 2019 Jie Cui et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

