Characterization of the c10orf76-PI4KB complex and its necessity for Golgi PI4P levels and enterovirus replication
- PMID: 31829496
- PMCID: PMC7001497
- DOI: 10.15252/embr.201948441
Characterization of the c10orf76-PI4KB complex and its necessity for Golgi PI4P levels and enterovirus replication
Abstract
The lipid kinase PI4KB, which generates phosphatidylinositol 4-phosphate (PI4P), is a key enzyme in regulating membrane transport and is also hijacked by multiple picornaviruses to mediate viral replication. PI4KB can interact with multiple protein binding partners, which are differentially manipulated by picornaviruses to facilitate replication. The protein c10orf76 is a PI4KB-associated protein that increases PI4P levels at the Golgi and is essential for the viral replication of specific enteroviruses. We used hydrogen-deuterium exchange mass spectrometry to characterize the c10orf76-PI4KB complex and reveal that binding is mediated by the kinase linker of PI4KB, with formation of the heterodimeric complex modulated by PKA-dependent phosphorylation. Complex-disrupting mutations demonstrate that PI4KB is required for membrane recruitment of c10orf76 to the Golgi, and that an intact c10orf76-PI4KB complex is required for the replication of c10orf76-dependent enteroviruses. Intriguingly, c10orf76 also contributed to proper Arf1 activation at the Golgi, providing a putative mechanism for the c10orf76-dependent increase in PI4P levels at the Golgi.
Keywords: GBF1; HDX-MS; PI4KB; c10orf76; viral replication.
© 2019 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Recombinant c10orf76 directly binds to PI4KB in vitro. His‐pulldown assays of baculovirus/Sf9 produced 6xHis‐c10orf76 (3 μM) were carried out with untagged PI4KB (2.5 μM).
PI4KB and c10orf76 form a stable complex. The complex of c10orf76‐PI4KB eluted from a S200 superdex 10/300 GL increase gel filtration column (GE) at a volume consistent with a heterodimer (169 kDa), while c10orf76 alone eluted at a volume consistent with a monomer (79 kDa). Lines with MW values indicate elution of MW standards (158 kDa aldolase, 75 kDa conalbumin).
PI4KB is potently inhibited by c10orf76 in a dose‐dependent manner in vitro. Kinase assays of PI4KB (20 nM) in the presence of varying concentrations of c10orf76 (1.6 nM–1 μM) were carried out on pure PI lipid vesicles (0.5 mg/l) in the presence of 100 μM ATP. The data were normalized to the kinase activity of PI4KB alone. IC50 values were determined by one binding site, nonlinear regression (curve fit) using Graphpad. Error bars represent standard deviation of independent technical replicates (n = 3).
PI4KB is potently inhibited by c10orf76 on pure PI vesicles and vesicles mimicking Golgi composition. Kinase assays of PI4KB and c10orf76 were carried out on lipid substrate composed of pure PI vesicles (0.5 mg/ml) with 100 μM ATP, and Golgi‐mimic vesicles (0.5 mg/ml, 10% PS, 20% PI, 25% PE, 45% PC) with 10 μM ATP. PI4KB was present at 20 and 300 nM in the PI and Golgi substrate assays, respectively, with c10orf76 present at 500 nM in both experiments. The data are normalized to the kinase activity of PI4KB alone. Error bars represent standard deviation of independent technical replicates (n = 3).
Changes in deuterium incorporation PI4KB in the presence of c10orf76 showed a profound ordering of the kinase domain N‐lobe linker and smaller changes in the helical domain and C‐lobe of the kinase domain. The sum of the difference mapped as the difference in number of deuterons incorporated for PI4KB (400 nM) in the presence and absence of c10orf76 (400 nM) over all time points (3 s at 1°C; 3 s, 30 s, and 300 s at 23°C). Each dot represents a peptide graphed on the x‐axis according to the central residue. The red boxes highlight key regions that showed significant changes (> 7% decrease in exchange, > 0.5 Da difference, and unpaired two‐tailed student t‐test P < 0.05). Error bars represent standard deviation of independent technical replicates (n = 3).
c10orf76 binding induces differences in HDX throughout multiple domains of PI4KB. Regions of > 7% difference in deuterium exchange in the presence of c10orf76 are colored onto the structure of PI4KB according to the legend (PDB: 4D0L). The N‐lobe linker of the kinase domain is disordered in the structure and is represented by a dotted line.
Changes in the deuterium incorporation of c10orf76 in the presence of PI4KB. H/D exchange reactions displayed as the sum of the difference in HDX in the number of deuterons for c10orf76 (400 nM) in the presence of PI4KB (400 nM) at all time points (3 s at 1°C; 3 s, 30 s, and 300 s at 23°C) analyzed. Red boxes highlight regions that showed significant changes (> 7% decrease in exchange, > 0.5 Da difference, and unpaired two‐tailed Student's t‐test P < 0.05). Error bars represent standard deviation of independent technical replicates (n = 3).
The PI4KB N‐lobe linker undergoes a disorder‐to‐order transition upon binding c10orf76. Selected peptides (including the sequence, domain information, and numbering) of both PI4KB and c10orf76 displayed as the % deuteration incorporation over time. Error bars represent standard deviation of independent technical replicates (n = 3) and are typically smaller than the size of the point on the graph.

PI4KB can form ternary complexes with Rab11a and c10orf76 in vitro. GST‐pulldown assays were carried out using GST‐Rab11a(Q70L) (6 μM) or GST alone (3 μM) as the bait, using 6xHis‐c10orf76 (4 μM), PI4KB (2 μM) as the prey. Samples were washed a total of four times in all experiments.
PI4KB can form ternary complexes with ACBD3 and c10orf76 in vitro. GST‐pulldown assays were carried out using GST‐ACBD3 (4 μM) or GST alone (4 μM) as the bait, and 6xHis‐c10orf76 (3 μM) and PI4KB (2 μM) as the prey. Samples were washed a total of four times in all experiments.

- A, B
All peptides of both PI4KB (A) and c10orf76 (B) with a significant difference in H/D exchange with > 7% decrease in exchange, > 0.5 Da difference, and unpaired two‐tailed Student's t‐test P < 0.05 at any time point (3 s at 1°C; 3 s, 30 s, and 300 s at 23°C). Error bars represent standard deviation of independent technical replicates (n = 3) and are typically smaller than the size of the point on the graph.

The N‐lobe kinase linker region of PI4KB is strongly conserved back to D. rerio. The N‐lobe linker region of PI4KB sequences of the organisms indicated was analyzed using Clustal Omega/ESpript 3. The consensus PKA motif (RRXS) that is conserved back to D. rerio is indicated on the sequence, as well as the RL494EA point mutation.
The N‐lobe kinase linker of PI4KB can be efficiently phosphorylated by PKA. Recombinant PKA at different concentrations (0, 7, 34, 168, or 840 ng) was incubated with recombinant (E. coli) wild‐type PI4KB (20 μg) for 1 h with 200 μM ATP, and the amount of phosphorylation was followed using mass spectrometry. Relative abundance of Ser496 phosphorylated PI4KB was calculated using the relative intensity (total area) of the phosphorylated vs non‐phosphorylated peptide (486–506).
PI4KB phosphorylation by PKA alters the affinity for c10orf76. The kinase activity of different variants of PI4KB (15 nM) was measured in the presence of varying amounts of c10orf76 (23 nM–1.5 μM) with 100% PI lipid substrate (0.5 mg/l) and 100 μM ATP. The data were normalized to the kinase activity of PI4KB alone. Error bars represent standard deviation of independent technical replicates (n = 3).
PI4KB has the same kinase activity when Ser496 is phosphorylated or mutated to alanine. Kinase assay of PI4KB non‐phosphorylated, phos‐Ser496, or S496A (15 nM) on pure PI lipid vesicles (0.5 mg/l) with 100 μM ATP. The data were normalized to the kinase activity of WT PI4KB. Error bars represent standard deviation of independent technical replicates (n = 3).
Engineered RL494EA PI4KB mutant shows decreased binding to c10orf76. His‐pulldown assays of 6xHis‐c10orf76 (3 μM) with wild‐type or RL494EA PI4KB (1–2 μM).
RL494EA PI4KB activity is not inhibited by c10orf76. Kinase assays of either wild‐type or mutant RL494EA PI4KB (40 nM) were carried out with varying concentrations of c10orf76 (3.9 nM–2 μM) with 100% PI lipid vesicles (0.5 mg/l) and 100 μM ATP. The data were normalized to the kinase activity of PI4KB alone. Error bars represent standard deviation of independent technical replicates (n = 3).
Wild‐type PI4KB and RL494EA PI4KB mutant have the same lipid kinase activity. Kinase assays of either wild‐type and mutant PI4KB (10 nM) were carried out with 100% PI lipid vesicles (0.5 mg/l), 100 μM ATP, and PI4KB (300 nM) on Golgi‐mimic vesicles (0.5 mg/ml) with 10 μM ATP. The data were normalized to the kinase activity of WT PI4KB. Error bars represent standard deviation of independent technical replicates (n = 3).
FLH409AAA‐c10orf76 mutant shows decreased affinity for PI4KB. His‐pulldown assays of 6xHis‐c10orf76 (1 μM) with wild‐type PI4KB (1 μM). Samples washed a total of four times.
Kinase assay shows FLH409AAA c10orf76 inhibition of PI4KB is greatly reduced. Kinase assay of PI4KB (40 nM) and a concentration curve of c10orf76 (3.9 nM–2 μM) on pure PI lipid vesicles (0.5 mg/l) with 100 μM ATP. The data were normalized to the kinase activity of PI4KB alone. Error bars represent standard deviation of independent technical replicates (n = 3).
The PI4KB‐binding region of c10orf76 is strongly conserved back to D. rerio. Clustal Omega/ESpript 3 alignment of the FLH409 region of c10orf76 that binds PI4KB.
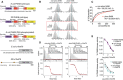
PKA phosphorylates the Ser496 site in PI4KB. Relative abundance of phosphorylated PI4KB at Ser294, Ser413, and Ser496 sites and c10orf76 at S14 and S/T from 325 to 351 expressed in Sf9, E. coli, or expressed in E. coli and treated with PKA (PI4KB) were calculated using the relative intensity (total area) of the phosphorylated vs non‐phosphorylated peptides (290–303, 290–312, 395–425, 430–441, 486–505, and 486–506) for PI4KB and (10–22 and 325–351) for c10orf76. For 325–351, the definitive phosphorylated Ser/Thr residue could not be determined. N.D. indicates no phosphorylation was identified.
Raw deuterium incorporation data for PI4KB peptide 488–508 used to generate panel (C). The deuterium incorporation for the phosphorylated and non‐phosphorylated variants of PI4KB is shown in the presence of different concentrations of c10orf76. Dotted blue line indicates mass centroid of PI4KB peptide in absence of c10orf76, and red box highlights the clear difference in deuterium incorporation at 40 nM c10orf76 between non‐phosphorylated and phosphorylated Ser496 PI4KB.
Phosphorylation of Ser496 reduces PI4KB affinity for c10orf76. Deuterium incorporation of the PI4KB kinase linker region peptide 488–508 (20 nM) at a single time point (5 s of D2O exposure at 23°C) was monitored in the presence of increasing concentrations of c10orf76 (0–320 nM c10orf76). Kd values were generated using a one binding site, nonlinear regression (curve fit), and are shown with 95% confidence intervals. Error bars represent standard deviation (n = 3), and most are smaller than the size of the point.
Representative ITC binding isotherms following the titration of c10orf76 into a solution of non‐phosphorylated PI4KB (left) or Ser496 phosphorylated PI4KB (right). Parameter values are an average with standard deviation of independent technical replicates (N = 3).
PI4KB S496D/E mutants do not mimic Ser496 phosphorylation modulation of c10orf76 binding. The kinase activity of S496D/E variants of PI4KB (15 nM) was measured in the presence of varying amounts of c10orf76 (23 nM–1.5 μM) with 100% PI lipid substrate (0.5 mg/l) and 100 μM ATP. The data were normalized to the kinase activity of PI4KB alone. Error bars represent standard deviation of independent technical replicates (n = 3).

Transfections of HEK293 cells revealed that both wild‐type GFP‐PI4KB and RL494EA GFP‐PI4KB localize to the Golgi.
WT c10orf76 also localized to the Golgi; however, the PI4KB binding‐deficient mutant of c10orf76 (FLH409AAA) predominantly localized to the cytosol.
Cartoon schematic of rapamycin‐inducible mitochondria recruitment. The AKAP1‐FRB‐CFP construct is localized to the outer mitochondrial membrane, while the mRFP‐FKBP12‐PI4KB and eGFP‐c10orf76 are localized in the Golgi as well as within the cytoplasm where they can form a complex. Upon addition of rapamycin, the mRFP‐FKBP12‐PI4KB construct is translocated to the mitochondria.
Mitochondria recruitment experiment with wild‐type PI4KB and c10orf76. Left: AKAP1‐FRB‐CFP is localized to the mitochondria before (top) and 5 min after rapamycin (100 nM) treatment (bottom). Middle: mRFP‐FKBP12‐PI4KB at high expression level is located in the cytosol before rapamycin (top) and translocates to the mitochondria after rapamycin induction (bottom). Right: eGFP‐c10orf76 at a high expression level is located in the cytosol before rapamycin (top) and translocates to the mitochondria after rapamycin induction (bottom).
Schematic of the rapamycin‐inducible mitochondria recruitment experiment with mutant PI4KB and WT c10orf76.
Mitochondria recruitment experiment with mutant PI4KB and WT c10orf76. Left: AKAP1‐FRB‐CFP is localized to the mitochondria before (top) and 5 min after (bottom) rapamycin treatment. Middle: mRFP‐FKBP12‐PI4KB(RL494EA) is located in the cytosol before rapamycin (top) and translocates to the mitochondria after rapamycin induction (bottom). Right: eGFP‐c10orf76 is located in the cytosol before (top) and after (bottom) rapamycin induction.
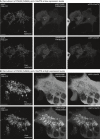
Mitochondria recruitment experiment with PI4KB(S496A) and c10orf76 at low expression levels. Left: AKAP1‐FRB‐CFP is localized to the mitochondria before (top) and 5 min after rapamycin (100 nM) treatment (bottom). Middle: mRFP‐FKBP12‐PI4KB(S496A) at a low expression level is located at the Golgi before rapamycin (top) and translocates to the mitochondria after rapamycin induction (bottom). Right: eGFP‐c10orf76 at a low expression level is located at the Golgi before rapamycin (top) and translocates to the mitochondria after rapamycin induction (bottom).
Mitochondria recruitment experiment with PI4KB(S496A) and c10orf76 at high expression levels (single cell zoom‐in). Left: AKAP1‐FRB‐CFP is localized to the mitochondria before (top) and 5 min after rapamycin (100 nM) treatment (bottom). Middle: mRFP‐FKBP12‐PI4KB(S496A) at a high expression level saturates the Golgi and cytosol before rapamycin (top) and robustly translocates to the mitochondria after rapamycin induction (bottom). Right: eGFP‐c10orf76 at a high expression level saturates the Golgi and cytosol before rapamycin (top) and robustly translocates to the mitochondria after rapamycin induction (bottom).
- A, B
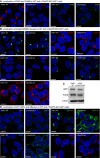
HAP1 cells were fixed and stained with antibodies examining PI4P and PI4KB (A) and Golgi morphology markers (B).
- C
Western blot of GBF1, PI4KB, and B‐Actin levels in both wild‐type and c10orf76 KO HAP1 cells.
- D
Markers of Arf1 activation—the coatomer proteins COPIα/γ and βCOP act as a readout for GTP‐bound Arf1, while the native coatomer was detected with the CM1 antibody.
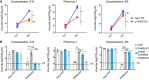
Viral infection assays determining viral titers by end‐point titration at 0 and 8 h in HAP1 wild‐type or c10orf76 knockout cells. Left: Coxsackievirus A10 infection. Middle: Poliovirus‐1 infection. Right: Coxsackievirus B3 infection. Titer values for c10orf76 knockout cells at 8 h were statistically evaluated compared to c10orf76 knockout titers at 0 h and wild‐type titers at 8 h using a one‐way ANOVA. **P < 0.01; N.S.: P > 0.05.
Viral infection assays determining virus titers by end‐point titration at 8 h in HeLa wild‐type and PI4KB knockout cells upon transfection of wild‐type PI4KB, the complex‐disrupting RL494EA PI4KB mutant or the kinase‐dead D674A PI4KB mutant. Left: Coxsackievirus A10 infection. Middle: Poliovirus‐1 infection. Right: Coxsackievirus B3 infection. Values were statistically evaluated compared to the GalT control using a one‐way ANOVA. **P < 0.01; *P < 0.05; N.S.: P > 0.05.

At the Golgi, the recruitment of PI4KB can be controlled through the action of ACBD3. PI4KB can recruit c10orf76 to the Golgi, with this playing a role in the dynamics and localization of GBF1 and active Arf1‐GTP and their downstream effectors (1). Active, membrane localized Arf1‐GTP can activate PI4KB (2).
In a c10orf76‐knockout cell, there is decreased recruitment of GBF1 and active Arf1, with this potentially describing the decreased levels of PI4P.
In enterovirus infected cells, there are multiple mechanisms that can alter PI4P levels and Arf1 activation. The 3A proteins from different viruses can activate PI4KB, primarily through hijacking ACBD3 (i), as well as directly binding to GBF1 (ii). Viruses that require c10orf76 to replicate rely on the PI4KB‐c10orf76 interface, emphasizing the importance of manipulating PI4KB‐c10orf76 and GBF1/Arf1‐GTP dynamics to facilitate viral replication.
Comment in
-
Another hijack! Some enteroviruses co-opt the c10orf76/PI4KB complex for their own good.EMBO Rep. 2020 Feb 5;21(2):e49876. doi: 10.15252/embr.201949876. Epub 2020 Jan 9. EMBO Rep. 2020. PMID: 31919962 Free PMC article.
References
-
- Tan J, Brill JA (2014) Cinderella story: PI4P goes from precursor to key signaling molecule. Crit Rev Biochem Mol Biol 49: 33–58 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- FRN 142393/Gouvernement du Canada|CIHR|Institute of Infection and Immunity (III)/International
- 17686/Michael Smith Foundation for Health Research (MSFHR)/International
- NWO-VICI-91812628/Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO)/International
- NWO-ECHO-725 711.017.002/Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO)/International
- 642434/EC|H2020|H2020 Priority Excellent Science|H2020 Marie Skłodowska-Curie Actions (MSCA)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

