Maintenance of active chromatin states by HMGN2 is required for stem cell identity in a pluripotent stem cell model
- PMID: 31831052
- PMCID: PMC6907237
- DOI: 10.1186/s13072-019-0320-7
Maintenance of active chromatin states by HMGN2 is required for stem cell identity in a pluripotent stem cell model
Abstract
Background: Members of the HMGN protein family modulate chromatin structure and influence epigenetic modifications. HMGN1 and HMGN2 are highly expressed during early development and in the neural stem/progenitor cells of the developing and adult brain. Here, we investigate whether HMGN proteins contribute to the chromatin plasticity and epigenetic regulation that is essential for maintaining pluripotency in stem cells.
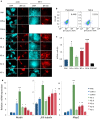
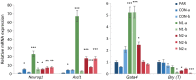
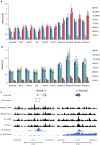
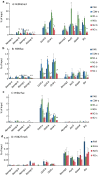
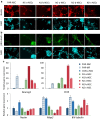
Results: We show that loss of Hmgn1 or Hmgn2 in pluripotent embryonal carcinoma cells leads to increased levels of spontaneous neuronal differentiation. This is accompanied by the loss of pluripotency markers Nanog and Ssea1, and increased expression of the pro-neural transcription factors Neurog1 and Ascl1. Neural stem cells derived from these Hmgn-knockout lines also show increased spontaneous neuronal differentiation and Neurog1 expression. The loss of HMGN2 leads to a global reduction in H3K9 acetylation, and disrupts the profile of H3K4me3, H3K9ac, H3K27ac and H3K122ac at the Nanog and Oct4 loci. At endodermal/mesodermal genes, Hmgn2-knockout cells show a switch from a bivalent to a repressive chromatin configuration. However, at neuronal lineage genes whose expression is increased, no epigenetic changes are observed and their bivalent states are retained following the loss of HMGN2.
Conclusions: We conclude that HMGN1 and HMGN2 maintain the identity of pluripotent embryonal carcinoma cells by optimising the pluripotency transcription factor network and protecting the cells from precocious differentiation. Our evidence suggests that HMGN2 regulates active and bivalent genes by promoting an epigenetic landscape of active histone modifications at promoters and enhancers.
Keywords: Chromatin; Differentiation; Embryonal carcinoma cells; Epigenetics; HMGN; Neuronal; Stem cells.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

