Systematic genome editing of the genes on zebrafish Chromosome 1 by CRISPR/Cas9
- PMID: 31831591
- PMCID: PMC6961580
- DOI: 10.1101/gr.248559.119
Systematic genome editing of the genes on zebrafish Chromosome 1 by CRISPR/Cas9
Abstract
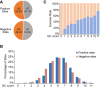
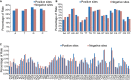
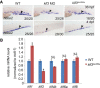
Genome editing by the well-established CRISPR/Cas9 technology has greatly facilitated our understanding of many biological processes. However, a complete whole-genome knockout for any species or model organism has rarely been achieved. Here, we performed a systematic knockout of all the genes (1333) on Chromosome 1 in zebrafish, successfully mutated 1029 genes, and generated 1039 germline-transmissible alleles corresponding to 636 genes. Meanwhile, by high-throughput bioinformatics analysis, we found that sequence features play pivotal roles in effective gRNA targeting at specific genes of interest, while the success rate of gene targeting positively correlates with GC content of the target sites. Moreover, we found that nearly one-fourth of all mutants are related to human diseases, and several representative CRISPR/Cas9-generated mutants are described here. Furthermore, we tried to identify the underlying mechanisms leading to distinct phenotypes between genetic mutants and antisense morpholino-mediated knockdown embryos. Altogether, this work has generated the first chromosome-wide collection of zebrafish genetic mutants by the CRISPR/Cas9 technology, which will serve as a valuable resource for the community, and our bioinformatics analysis also provides some useful guidance to design gene-specific gRNAs for successful gene editing.
© 2020 Sun et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
