Functional interplay between protein domains in a supramodular structure involving the postsynaptic density protein PSD-95
- PMID: 31831623
- PMCID: PMC7029118
- DOI: 10.1074/jbc.RA119.011050
Functional interplay between protein domains in a supramodular structure involving the postsynaptic density protein PSD-95
Abstract
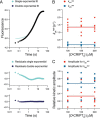
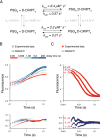
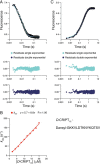
Cell scaffolding and signaling are governed by protein-protein interactions. Although a particular interaction is often defined by two specific domains binding to each other, this interaction often occurs in the context of other domains in multidomain proteins. How such adjacent domains form supertertiary structures and modulate protein-protein interactions has only recently been addressed and is incompletely understood. The postsynaptic density protein PSD-95 contains a three-domain supramodule, denoted PSG, which consists of PDZ, Src homology 3 (SH3), and guanylate kinase-like domains. The PDZ domain binds to the C terminus of its proposed natural ligand, CXXC repeat-containing interactor of PDZ3 domain (CRIPT), and results from previous experiments using only the isolated PDZ domain are consistent with the simplest scenario for a protein-protein interaction; namely, a two-state mechanism. Here we analyzed the binding kinetics of the PSG supramodule with CRIPT. We show that PSG binds CRIPT via a more complex mechanism involving two conformational states interconverting on the second timescale. Both conformational states bound a CRIPT peptide with similar affinities but with different rates, and the distribution of the two conformational states was slightly shifted upon CRIPT binding. Our results are consistent with recent structural findings of conformational changes in PSD-95 and demonstrate how conformational transitions in supertertiary structures can shape the ligand-binding energy landscape and modulate protein-protein interactions.
Keywords: CXXC repeat–containing interactor of PDZ3 domain (CRIPT); PDZ domain; PSD-95; conformational change; kinetics; postsynaptic density protein; protein conformation; protein–protein interaction; supertertiary structure; supramodule.
© 2020 Laursen et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

