Control of noncoding RNA production and histone levels by a 5' tRNA fragment
- PMID: 31831626
- PMCID: PMC6938667
- DOI: 10.1101/gad.332783.119
Control of noncoding RNA production and histone levels by a 5' tRNA fragment
Erratum in
-
Corrigendum: Control of noncoding RNA production and histone levels by a 5' tRNA fragment.Genes Dev. 2020 Mar 1;34(5-6):462. doi: 10.1101/gad.336958.120. Genes Dev. 2020. PMID: 32122968 Free PMC article. No abstract available.
Abstract
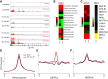
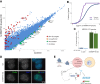
Small RNAs derived from mature tRNAs, referred to as tRNA fragments or "tRFs," are an emerging class of regulatory RNAs with poorly understood functions. We recently identified a role for one specific tRF-5' tRF-Gly-GCC, or tRF-GG-as a repressor of genes associated with the endogenous retroelement MERVL, but the mechanistic basis for this regulation was unknown. Here, we show that tRF-GG plays a role in production of a wide variety of noncoding RNAs-snoRNAs, scaRNAs, and snRNAs-that are dependent on Cajal bodies for stability and activity. Among these noncoding RNAs, regulation of the U7 snRNA by tRF-GG modulates heterochromatin-mediated transcriptional repression of MERVL elements by supporting an adequate supply of histone proteins. Importantly, the effects of inhibiting tRF-GG on histone mRNA levels, on activity of a histone 3' UTR reporter, and ultimately on MERVL regulation could all be suppressed by manipulating U7 RNA levels. We additionally show that the related RNA-binding proteins hnRNPF and hnRNPH bind directly to tRF-GG, and are required for Cajal body biogenesis, positioning these proteins as strong candidates for effectors of tRF-GG function in vivo. Together, our data reveal a conserved mechanism for 5' tRNA fragment control of noncoding RNA biogenesis and, consequently, global chromatin organization.
Keywords: epigenetics; histones; tRNA fragment.
© 2020 Boskovic et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
