Development of a membrane lipid metabolism-based signature to predict overall survival for personalized medicine in ccRCC patients
- PMID: 31832113
- PMCID: PMC6882998
- DOI: 10.1007/s13167-019-00189-8
Development of a membrane lipid metabolism-based signature to predict overall survival for personalized medicine in ccRCC patients
Abstract
Background: Clear cell renal cell carcinoma (ccRCC) is the most common type of renal cell carcinoma and is characterized by a dysregulation of changes in cellular metabolism. Altered lipid metabolism contributes to ccRCC progression and malignancy.
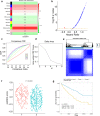
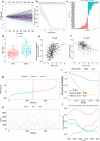
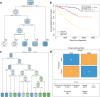
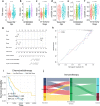
Method: Associations among survival potential and each gene ontology (GO) term were analyzed by univariate Cox regression. The results revealed that membrane lipid metabolism had the greatest hazard ratio (HR). Weighted gene co-expression network analysis (WGCNA) was applied to determine the key genes associated with membrane lipid metabolism. Consensus clustering was used to identify novel molecular subtypes based on the key genes. LASSO Cox regression was performed to build a membrane lipid metabolism-based signature. The random forest algorithm was applied to find the most important mutations associated with membrane lipid metabolism. Decision trees and nomograms were constructed to quantify risks for individual patients.
Result: Membrane lipid metabolism stratified ccRCC patients into high- and low-risk groups. Key genes were identified by WGCNA. Membrane lipid metabolism-based signatures exhibited higher prediction efficiency than other clinicopathological traits in both whole cohort and subgroup analyses. The random forest algorithm revealed high associations among the membrane lipid metabolism-based signature and BAP1, PBRM1 and VHL mutations. Decision trees and nomograms indicated high efficiency for risk stratification.
Conclusion: Our study might contribute to the optimization of risk stratification for survival and personalized management of ccRCC patients.
Keywords: Algorithm; Clear cell renal cell carcinoma (ccRCC); Decision tree; Gene co-expression network analysis; Gene signature; Membrane lipid metabolism; Overall survival; Patient stratification; Predictive preventive personalized medicine (PPPM); Risk assessment; Somatic mutations; von Hippel-Lindau (VHL).
© European Association for Predictive, Preventive and Personalised Medicine (EPMA) 2019.
Conflict of interest statement
Conflict of interestThe authors declare that they have no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

