Morphological and stage-specific transcriptome analyses reveal distinct regulatory programs underlying yam (Dioscorea alata L.) bulbil growth
- PMID: 31832647
- PMCID: PMC7242083
- DOI: 10.1093/jxb/erz552
Morphological and stage-specific transcriptome analyses reveal distinct regulatory programs underlying yam (Dioscorea alata L.) bulbil growth
Abstract
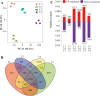
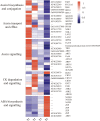
In yam (Dioscorea spp) species, bulbils at leaf axils are the most striking species-specific axillary structure and exhibit important ecological niches. Genetic regulation underlying bulbil growth remains largely unclear so far. Here, we characterize yam (Dioscorea alata L.) bulbil development using histological analysis, and perform full transcriptional profiling on key developmental stages together with phytohormone analyses. Using the stage-specific scoring algorithm, we have identified 3451 stage-specifically expressed genes that exhibit a tight link between major transcriptional changes and stages. Co-expressed gene clusters revealed an obvious over-representation of genes associated with cell division and expansion at the initiation stage of bulbils (T1). Transcriptional changes of hormone-related genes highly coincided with hormone levels, indicating that bulbil initiation and growth are coordinately controlled by multiple phytohormones. In particular, localized auxin is transiently required to trigger bulbil initiation, and be further depleted or exported from bulbils to promote growth by up-regulation of genes involved in auxinconjugation and efflux. The sharp increase in supply of sucrose and an enhanced trehalose-6-phophate pathway at T1 were observed, suggesting that sucrose probably functions as a key signal and promotes bulbil initiation. Analysis of the expression of transcription factors (TFs) predicated 149 TFs as stage-specifically expressed; several T1-specific TFs (from Aux/IAA, E2F, MYB, and bHLH families) have been shown to play key roles in triggering bulbil formation. Together, our work provides a crucial angle for in-depth understanding of the molecular programs underlying yam's unique bulbil development processes. Stage-specific gene sets can be queried to obtain key candidates regulating bulbil growth, serving as valuable resources for further functional research.
Keywords: Bulbil; genetic regulation; growth; phytohormone; transcriptome; yam (Dioscorea alata L).
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Experimental Biology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures






References
-
- Abraham-Juárez MJ, Martínez-Hernández A, Leyva-González MA, Herrera-Estrella L, Simpson J. 2010. Class I KNOX genes are associated with organogenesis during bulbil formation in Agave tequilana. Journal of Experimental Botany 61, 4055–4067. - PubMed
-
- Alexa A, Rahnenführer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607. - PubMed
-
- Arizaga S, Ezcurra E. 2002. Propagation mechanisms in Agave macroacantha (Agavaceae), a tropical arid-land succulent rosette. American Journal of Botany 89, 632–641. - PubMed
-
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. 2005. Cytokinin oxidase regulates rice grain production. Science 309, 741–745. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

