Armitage determines Piwi-piRISC processing from precursor formation and quality control to inter-organelle translocation
- PMID: 31833223
- PMCID: PMC7001504
- DOI: 10.15252/embr.201948769
Armitage determines Piwi-piRISC processing from precursor formation and quality control to inter-organelle translocation
Abstract
Piwi and piRNA form the piRNA-induced silencing complex (piRISC) to repress transposons. In the current model, Armitage (Armi) brings the Piwi-piRISC precursor (pre-piRISC) to mitochondria, where Zucchini-dependent piRISC maturation occurs. Here, we show that Armi is necessary for Piwi-pre-piRISC formation at Yb bodies and that Armi triggers the exit of Piwi-pre-piRISC from Yb bodies and the translocation to mitochondria. Piwi-pre-piRISC resist leaving Yb bodies until Armi binds Piwi-pre-piRISC through the piRNA precursors. The lack of the Armi N-terminus also blocks the Piwi-pre-piRISC exit from Yb bodies. Thus, Armi determines Piwi-piRISC processing, in a multilayered manner, from precursor formation and quality control to inter-organelle translocation for maturation.
Keywords: Armitage; Gasz; Yb body; mitochondrion; piRNA.
© 2019 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Western blotting using anti‐Gasz monoclonal antibody shows that the Gasz signal disappeared upon Gasz RNAi treatment in OSCs. Anti‐Gasz monoclonal antibody was produced in this study. β‐Tubulin (β‐Tub) was detected as a loading control.
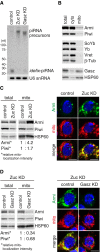
Western blotting shows that the levels of Armi and Piwi in the cytoplasmic fraction after mitochondrial isolation (cyto) were little impacted by Zuc depletion, while the level of Armi in the mitochondrial fraction (mito) increased.
Larger immunostaining views of the cell images shown in Fig 1C and D. The cells appeared in the main figures are boxed with white dotted line. In “Zuc KD” panel, the upper cell appeared in Fig 1C, while the lower one appeared in Fig 1D. Armi behaved similarly in the cells other than the one shown in Fig 1C and D. The scale bar represents 5 μm.
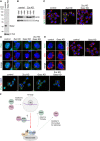
Upper: Localization of Piwi (green) in OSCs upon Zuc and Gasz depletion. Yb bodies are shown in red. Lower: Larger immunostaining views of the cell images shown in upper panels. The cells appeared in the upper panels are boxed with white dotted line. Piwi was mislocalized in the cytosol upon the loss of Zuc and/or Gasz. Some fraction of Piwi was detected at Yb bodies as has previously been reported 24. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
Left: Localization of Armi (green) in OSCs upon Gasz depletion. Mitochondria are shown in red. Right: Larger immunostaining views of the cell images shown on the left. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
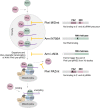
Piwi localizes to Yb bodies after Armi has localized there. Upon Piwi–piRISC formation by Zuc on the outer membrane of mitochondria, Armi goes back to Yb bodies, while Piwi–piRISC is translocated to the nucleus for silencing. The details and requirements of inter‐organelle translocation of Armi and Piwi–pre‐piRISC remain unknown (red dotted line with an arrowhead).
Northern blotting shows that Gasz depletion (Gasz KD) led to the accumulation of flam‐piRNA precursors but reduced the level of mature idefix‐piRNA arising from the precursors, as in the case of Zuc depletion (Zuc KD). U6 snRNA was detected as a loading control.
Western blotting showing the protein levels of endogenous Armi, Piwi, SoYb, Yb, Vret, and Gasz in total OSC lysate (total), the mitochondrial fraction (mito), and the cytoplasmic fraction after mitochondrial isolation (cyto). β‐Tubulin (β‐Tub) and HSP60 were detected as markers for the “cyto” and “mito” fractions, respectively.
Left: Western blotting comparing the abundances of Armi and Piwi in total OSC lysate (total) and the mitochondrial fraction (mito) before (control) and after (Zuc KD) Zuc knockdown in OSCs. Relative mito‐localization intensity (*) shows the amount of proteins in “mito” normalized by the amount of proteins in “total” and HSP60. Right: Immunofluorescence shows that the Armi signal (green) is strongly overlapped with the mitochondrial signal (red) in Zuc‐depleted OSCs (Zuc KD). Armi is localized to Yb bodies in control OSCs (control). The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
Left: Western blotting comparing the abundances of Armi and Piwi in total OSC lysate (total) and the mitochondrial fraction (mito) before (control) and after (Gasz KD) Gasz depletion in Zuc‐depleted OSCs (Zuc KD). Relative mito‐localization intensity (*) shows the amount of proteins in “mito” normalized by the amount of proteins in “total” and HSP60. Right: Immunofluorescence shows that the Armi mitochondrial signal (green) found in Zuc‐depleted cells (Zuc KD/control) mostly disappeared after additional Gasz depletion in the cells (Zuc KD/Gasz KD), but was found at Yb bodies. Mitochondrial signal is shown in red. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
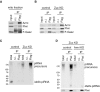
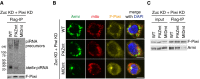
Armi and Piwi co‐immunoprecipitated with Flag‐Gasz (F‐Gasz) from the mitochondrial fraction (mito fraction) of OSCs (input). F‐Gasz was ectopically expressed in the cells by transfection prior to immunoprecipitation. *: Heavy chains of antibodies used for immunoprecipitation. n.i.: Non‐immune IgG used as a negative control.
Co‐immunoprecipitation from whole‐cell lysate of OSCs before (control) and after (Zuc KD) Zuc depletion and subsequent Western blotting show that the levels of Piwi and Armi in the Flag‐Gasz (F‐Gasz) complex increased upon Zuc depletion (Zuc KD). Armi signal with a longer exposure is shown in Fig EV2A.
Northern blotting shows that the flam‐piRNA precursors in the Flag‐Gasz complex (Flag) accumulated upon Zuc depletion (Zuc KD). A gel image of the control lanes (control) with a long exposure is shown in Fig EV2B.
Upper: The flam‐piRNA precursors associated with Piwi disappeared upon Armi depletion (Armi KD) in Zuc‐lacking OSCs (Zuc KD). Lower: The abundance of immunopurified Piwi is shown by Western blotting.

This Western blot is identical to the blot in Fig 2B (Armi) but with a longer exposure time.
This northern blot is identical to the blot in Fig 2C (left half) but with a longer exposure time.

- A, B
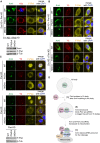
Depletion of endogenous Piwi (Piwi KD) in Zuc‐lacking OSCs (Zuc KD) caused Armi (green) to relocate to Yb bodies. Localization of Armi at Yb bodies is shown in (B). Mitochondrial (upper panel) and Yb (lower panel) signals are shown in red. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
- B
The Armi complex was immunopurified from Zuc‐lacking OSCs (Zuc KD) before (control) and after (Piwi KD) Piwi depletion and probed for detecting Armi and Gasz. Armi no longer stably associated with endogenous Gasz upon Piwi depletion in Zuc‐lacking OSCs. Co‐IP index (*) shows that the ratio of Gasz amount in the Armi complex isolated from Zuc‐depleted cells (Zuc KD/control) and Zuc‐ and Piwi‐depleted cells (Zuc KD/Piwi KD) is 1:0.05.
- C
The Flag‐Gasz (F‐Gasz) complex was immunopurified from Zuc‐lacking OSCs (Zuc KD) before (control) and after (Piwi KD) Piwi depletion and probed for detecting Armi and F‐Gasz. F‐Gasz was ectopically expressed prior to immunoprecipitation. Co‐IP index (*) shows that the ratio of Armi amount in the F‐Gasz complex isolated from Zuc‐depleted cells (Zuc KD/control) and Zuc‐ and Piwi‐depleted cells (Zuc KD/Piwi KD) is 1:0.31.
- D
In vitro pull‐down assays show that Armi‐Flag (Armi‐F) directly binds with GST‐GaszΔC113 but hardly with GST. Armi‐F was immunopurified from Schneider 2 (S2) cells under harsh conditions. GST and GST‐GaszΔC113 were visualized by CBB staining, while Armi‐F was detected by Western blotting using anti‐Flag antibodies.
Larger immunostaining views of the cell images shown in Fig 3A and B. The cells appeared in the main figures are boxed with white dotted line. Armi behaved similarly in the cells other than the one shown in Fig 3A and B. The scale bar represents 5 μm.
Piwi depletion hardly affected the level of Armi in Zuc‐depleted OSCs. β‐Tubulin (β‐Tub) was detected as a loading control.
Myc‐Gasz (M‐Gasz) and Armi‐Flag (Armi‐F) interacted with each other in Schneider 2 (S2) cells.
Purified recombinant GST‐Gasz (ΔC113 lacking Leu349‐Ser461 containing the TM region at the C‐terminus) used in the experiment shown in Fig 3E. The protein was stained with CBB.
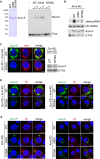
Upper: Northern blotting shows that Flag‐Piwi WT (WT) and PAZ mutant (PAZmt) but not MID mutant (MIDmt) bound with flam‐piRNA precursors in Zuc‐depleted OSCs. Endogenous Piwi was depleted simultaneously with Zuc (Zuc KD + Piwi KD). Zuc was depleted by RNAi and so residual Zuc may have remained in cells. Therefore, a tiny amount of piRNAs was loaded onto Piwi. F‐Piwi WT, PAZmt, and MIDmt were made to be RNAi‐resistant by mutagenesis (see Materials and Methods). Lower: The abundance of immunopurified Flag‐Piwi (F‐Piwi) is shown by Western blotting.
Armi localization to Yb bodies observed in Zuc‐ and Piwi‐depleted OSCs (Zuc KD + Piwi KD) (Fig 3A and B) was restored by ectopic expression of Flag‐Piwi WT and PAZmt but not of MIDmt (see also Fig EV4B). Armi, mitochondria, and F‐Piwi are shown in green, red, and yellow, respectively. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
Western blotting shows that Flag‐Piwi (F‐Piwi) WT (WT) and PAZ mutant (PAZmt) but not MID mutant (MIDmt) bound with Armi in Zuc/Piwi‐depleted OSCs (Zuc KD + Piwi KD).
Upper: Another set of cell images of the one shown in Fig 4B. The scale bar represents 5 μm. DAPI (blue) shows the nuclei. Lower: The expression levels of Flag‐Piwi (F‐Piwi) WT and PAZ/MID mutants (PAZmt/MIDmt) in Zuc/Piwi‐depleted OSCs (Zuc KD + Piwi KD). β‐Tubulin (β‐Tub) was detected as a loading control.
Upper: Armi localization to Yb bodies observed in Zuc/Piwi‐depleted OSCs (Zuc KD + Piwi KD) (Fig 3A and B) was rescued by ectopic expression of Flag‐Piwi WT and PAZmt but not of MIDmt. “Rescued” means that Armi is now localized to mitochondria as in Zuc‐depleted OSCs (Fig 1C). The scale bar represents 5 μm. DAPI (blue) shows the nuclei. Lower: Another set of cell images of the one shown on the left. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
Upper: The cellular localization of Flag‐Piwi (F‐Piwi) WT and PAZ/MID mutants (PAZmt/MIDmt) (yellow), Yb (red), and Armi (green) in Piwi‐depleted OSCs (Piwi KD). Middle: Another set of cell images of the one shown on the left. The scale bar represents 5 μm. DAPI (blue) shows the nuclei. Lower: The expression levels of Flag‐Piwi (F‐Piwi) WT and PAZ/MID mutants (PAZmt/MIDmt) in Piwi‐depleted OSCs (Piwi KD). β‐Tubulin (β‐Tub) was detected as a loading control.
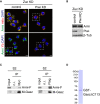
Armi has three functions. [A] Armi's Yb‐body localization is required for Piwi localization at Yb bodies. [B] Armi senses and binds Piwi–pre‐piRISC at Yb bodies. This action of Armi is required for inter‐organelle translocation of the complex from Yb bodies to mitochondria. [C] Armi relaxes piRNA precursors for Zuc cleavage on the mitochondria.
Left: Purified recombinant Armi‐Flag (Armi‐F) WT and mutants (ΔN34 and N756A) used in the experiment. Right: RNA‐binding activity of Armi‐Flag WT, ΔN34, and N756A. Here, 16‐nt single‐stranded RNA was 5′‐end‐labeled and incubated with Armi‐Flag.
Top: Northern blotting shows that siRNA‐resistant WT Armi‐Flag (Armi‐F), but not ΔN34 and N756A mutants, rescued the defects in idefix‐piRNA production caused by endogenous Armi depletion. U6 snRNA was used as a loading control. Bottom: The expression levels of Armi‐Flag (Armi‐F) WT, ΔN34, and N756A mutants. β‐Tubulin was used as a loading control.
Left: Another set of cell images of the one shown in Fig 5B. The scale bar represents 5 μm. DAPI (blue) shows the nuclei. Right: The expression levels of Armi‐Flag (Armi‐F) WT and N756A mutant in Zuc/Armi‐depleted OSCs (Zuc KD + Armi KD). β‐Tubulin (β‐Tub) was detected as a loading control.
Left: B. Armi‐Flag (Armi‐F) WT and the N756A mutant were expressed in OSCs where endogenous Armi and Zuc had been depleted by RNAi (Zuc KD + Armi KD). Armi‐F WT (green) localized onto mitochondria, whereas Armi‐F N756A mutant (green) localized to Yb bodies (red). The scale bar represents 5 μm. DAPI (blue) shows the nuclei. Right: Another set of cell images of the one shown on the left. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
Left: Armi‐Flag (Armi‐F) WT and ΔN34/N756A mutants (green) localized at Yb bodies (red) in Armi‐depleted OSCs (Armi KD). Right: Another set of cell images of the one shown on the left. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
Armi‐Flag (Armi‐F) WT and the N756A mutant were expressed in OSCs where endogenous Armi and Zuc had been depleted by RNAi (Zuc KD + Armi KD). Immunoprecipitation and subsequent Western blotting show that Armi‐F WT bound with Piwi and Gasz. The Armi N756A mutant bound with Piwi strongly but only weakly with Gasz. Luciferase‐Flag (Luc‐F) was used as a negative control.
Armi‐Flag (Armi‐F) WT and the N756A mutant were expressed in OSCs where endogenous Armi and Zuc had been depleted by RNAi (Zuc KD + Armi KD). Armi‐F WT (green) localized onto mitochondria (red), whereas Armi‐F N756A mutant (green) localized to Yb bodies (see also Fig EV5D). The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
In vitro pull‐down assays show that both Armi‐Flag (Armi‐F) WT and N756A mutant directly bind with GST‐GaszΔC113 but not with GST. Armi‐F WT and mutant were immunopurified from Schneider 2 (S2) cells under harsh conditions. GST and GST‐GaszΔC113 were visualized by CBB staining, while Armi‐F WT and N756A mutant were detected by Western blotting using anti‐Flag antibodies.

Anti‐Armi antibody 2F8A9 recognized both Armi bands on Western blots, while 2D6E11 recognized solely the upper band. The signals of 2F8A9 were stripped away, and then, the same membrane was reprobed with 2D6E11. β‐Tubulin (β‐Tub) was detected as a loading control.
Western blotting showed that 2F8A9 anti‐Armi antibody recognized both Armi‐Flag (Armi‐F) WT and ΔN34 mutant, while 2D6E11 recognized only WT. β‐Tubulin (β‐Tub) was detected as a loading control.
Left: Another set of cell images of the one shown in Fig 6B. The scale bar represents 5 μm. Right: The expression levels of Armi‐Flag (Armi‐F) WT and ΔN34 mutants in Zuc/Armi‐depleted OSCs (Zuc KD + Armi KD). β‐Tubulin (β‐Tub) was detected as a loading control.
Left: Armi‐Flag (Armi‐F) WT and the ΔN34 mutant were expressed in OSCs where endogenous Armi and Zuc had been depleted by RNAi (Zuc KD + Armi KD). Armi‐F WT (green) localized onto mitochondria, whereas Armi‐F ΔN34 mutant (green) localized to Yb bodies (red). The scale bar represents 5 μm. DAPI (blue) shows the nuclei. Right: Another set of cell images of the one shown on the left. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
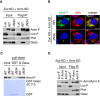
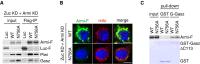
Armi‐Flag (Armi‐F) WT and the ΔN34 mutant were expressed in OSCs where endogenous Armi and Zuc had been depleted by RNAi (Zuc KD + Armi KD). Immunoprecipitation and subsequent Western blotting show that Armi‐F WT but not Armi ΔN34 mutant bound with Gasz in the cells. Both Armi WT and ΔN34 mutant interacted with Piwi to similar extents. Luciferase‐Flag (Luc‐F) was used as a negative control.
Armi‐Flag (Armi‐F) WT and the ΔN34 mutant were expressed in OSCs where endogenous Armi and Zuc had been depleted by RNAi (Zuc KD + Armi KD). Armi‐F WT (green) localized onto mitochondria (red), whereas Armi‐F ΔN34 mutant (green) localized to Yb bodies (see also Fig EV6D). The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
In vitro pull‐down assays show that both Armi‐Flag (Armi‐F) WT and ΔN34 mutant directly bind with GST‐GaszΔC113 but not with GST. Armi‐F WT and mutant were immunopurified from Schneider 2 (S2) cells under harsh conditions. GST and GST‐GaszΔC113 were visualized by CBB staining, while Armi‐F WT and mutant were detected by Western blotting using anti‐Flag antibodies.
Immunoprecipitation and subsequent Western blotting show that Armi‐Flag (Armi‐F) but not Flag‐Armi (F‐Armi) strongly bound with Gasz in Zuc/Armi‐depleted OSCs (Zuc KD + Armi KD). Piwi bound with both Armi (F‐Armi and Armi‐F). Luciferase‐Flag (Luc‐F) was used as a negative control.
References
-
- Ozata DM, Gainetdinov I, Zoch A, O'Carroll D, Zamore PD (2019) PIWI‐interacting RNAs: small RNAs with big functions. Nat Rev Genet 20: 89–108 - PubMed
-
- Yamashiro H, Siomi MC (2018) PIWI‐interacting RNA in Drosophila: biogenesis, transposon regulation, and beyond. Chem Rev 118: 4404–4421 - PubMed
-
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA (2001) Double‐stranded RNA‐mediated silencing of genomic tandem repeats and transposable elements in the melanogaster melanogaster germline. Curr Biol 11: 1017–1027 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

