Gene-environment regulatory circuits of right ventricular pathology in tetralogy of fallot
- PMID: 31834445
- PMCID: PMC7942233
- DOI: 10.1007/s00109-019-01857-y
Gene-environment regulatory circuits of right ventricular pathology in tetralogy of fallot
Abstract
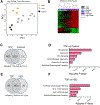
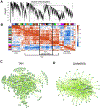
The phenotypic spectrum of congenital heart defects (CHDs) is contributed by both genetic and environmental factors. Their interactions are profoundly heterogeneous but may operate on common pathways as in the case of hypoxia signaling during postnatal heart development in the context of CHDs. Tetralogy of Fallot (TOF) is the most common cyanotic (hypoxemic) CHD. However, how the hypoxic environment contributes to TOF pathogenesis after birth is poorly understood. We performed Genome-wide transcriptome analysis on right ventricle outflow tract (RVOT) specimens from cyanotic and noncyanotic TOF. Co-expression network analysis identified gene modules specifically associated with clinical diagnosis and hypoxemia status in the TOF hearts. In particular, hypoxia-dependent induction of myocyte proliferation is associated with E2F1-mediated cell cycle regulation and repression of the WNT11-RB1 axis. Genes enriched in epithelial mesenchymal transition (EMT), fibrosis, and sarcomere were also repressed in cyanotic TOF patients. Importantly, transcription factor analysis of the hypoxia-regulated modules suggested CREB1 as a putative regulator of hypoxia/WNT11-RB1 circuit. The study provides a high-resolution landscape of transcriptome programming associated with TOF phenotypes and unveiled hypoxia-induced regulatory circuit in cyanotic TOF. Hypoxia-induced cardiomyocyte proliferation involves negative modulation of CREB1 activity upstream of the WNT11-RB1 axis. KEY MESSAGES: Genetic and environmental factors contribute to congenital heart defects (CHDs). How hypoxia contributes to Tetralogy of Fallot (TOF) pathogenesis after birth is unclear. Systems biology-based analysis revealed distinct molecular signature in CHDs. Gene expression modules specifically associated with cyanotic TOF were uncovered. Key regulatory circuits induced by hypoxia in TOF pathogenesis after birth were unveiled.
Keywords: Congenital heart defects; Genome; Hypoxia; Tetralogy of Fallot; Transcriptome.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

