A mechanical toy model linking cell-substrate adhesion to multiple cellular migratory responses
- PMID: 31834551
- PMCID: PMC6917673
- DOI: 10.1007/s10867-019-09536-2
A mechanical toy model linking cell-substrate adhesion to multiple cellular migratory responses
Abstract
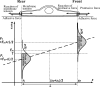
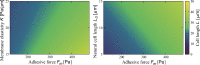
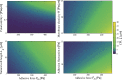
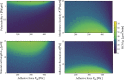
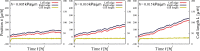
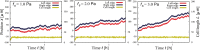
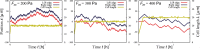
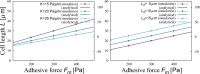
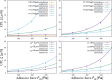
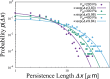
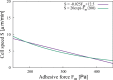
During cell migration, forces applied to a cell from its environment influence the motion. When the cell is placed on a substrate, such a force is provided by the cell-substrate adhesion. Modulation of adhesivity, often performed by the modulation of the substrate stiffness, tends to cause common responses for cell spreading, cell speed, persistence, and random motility coefficient. Although the reasons for the response of cell spreading and cell speed have been suggested, other responses are not well understood. In this study, we develop a simple toy model for cell migration driven by the relation of two forces: the adhesive force and the plasma membrane tension. The simplicity of the model allows us to perform the calculation not only numerically but also analytically, and the analysis provides formulas directly relating the adhesivity to cell spreading, persistence, and the random motility coefficient. Accordingly, the results offer a unified picture on the causal relations between those multiple cellular responses. In addition, cellular properties that would influence the migratory behavior are suggested.
Keywords: Cell migration; Chemokinesis; Modeling; Persistence; Random motility coefficient.
Conflict of interest statement
The author declares that he has no conflict of interest.
Figures
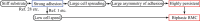
Similar articles
-
The matrix environmental and cell mechanical properties regulate cell migration and contribute to the invasive phenotype of cancer cells.Rep Prog Phys. 2019 Jun;82(6):064602. doi: 10.1088/1361-6633/ab1628. Epub 2019 Apr 4. Rep Prog Phys. 2019. PMID: 30947151 Review.
-
Interplay between substrate rigidity and tissue fluidity regulates cell monolayer spreading.Soft Matter. 2022 Oct 19;18(40):7877-7886. doi: 10.1039/d2sm00757f. Soft Matter. 2022. PMID: 36205535 Free PMC article.
-
A stochastic model for adhesion-mediated cell random motility and haptotaxis.J Math Biol. 1993;31(6):563-600. doi: 10.1007/BF00161199. J Math Biol. 1993. PMID: 8376918
-
A multiscale theory for spreading and migration of adhesion-reinforced mesenchymal cells.J R Soc Interface. 2023 Dec;20(209):20230317. doi: 10.1098/rsif.2023.0317. Epub 2023 Dec 13. J R Soc Interface. 2023. PMID: 38086406 Free PMC article.
-
Forces on adhesive contacts affect cell function.Curr Opin Cell Biol. 1998 Oct;10(5):566-71. doi: 10.1016/s0955-0674(98)80030-6. Curr Opin Cell Biol. 1998. PMID: 9818165 Review.
References
-
- Wang HB, Dembo M, Wang YL. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am. J. Physiol. Cell Physiol. 2000;279:C1345–C1350. - PubMed
-
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

