Toward a Common Coordinate Framework for the Human Body
- PMID: 31835027
- PMCID: PMC6934046
- DOI: 10.1016/j.cell.2019.11.019
Toward a Common Coordinate Framework for the Human Body
Abstract
Understanding the genetic and molecular drivers of phenotypic heterogeneity across individuals is central to biology. As new technologies enable fine-grained and spatially resolved molecular profiling, we need new computational approaches to integrate data from the same organ across different individuals into a consistent reference and to construct maps of molecular and cellular organization at histological and anatomical scales. Here, we review previous efforts and discuss challenges involved in establishing such a common coordinate framework, the underlying map of tissues and organs. We focus on strategies to handle anatomical variation across individuals and highlight the need for new technologies and analytical methods spanning multiple hierarchical scales of spatial resolution.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest
AR is a founder and equity holder of Celsius Therapeutics and an SAB member of ThermoFisher Scientific, Neogene Therapeutics, and Syros Pharmaceuticals.
Figures
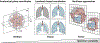
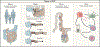


References
-
- Achim K et al. (2015) ‘High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin’, Nature biotechnology. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved, 33(5), pp. 503–509. - PubMed
-
- Aizarani N et al. (2019) ‘A human liver cell atlas reveals heterogeneity and epithelial progenitors’, Nature. nature.com, 572(7768), pp. 199–204. - PMC - PubMed
-
- Allassonnière S, Amit Y and Trouvé A (2007) ‘Towards a coherent statistical framework for dense deformable template estimation’, Journal of the Royal Statistical Society. Series B, Statistical methodology. Wiley Online Library, 69(1). doi: 10.1111/j.1467-9868.2007.00574.x. - DOI
-
- Allen Institute (2017) Allen Mouse Common Coordinate Framework. v3.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

