A Translation-Activating Function of MIWI/piRNA during Mouse Spermiogenesis
- PMID: 31835033
- PMCID: PMC8139323
- DOI: 10.1016/j.cell.2019.11.022
A Translation-Activating Function of MIWI/piRNA during Mouse Spermiogenesis
Abstract
Spermiogenesis is a highly orchestrated developmental process during which chromatin condensation decouples transcription from translation. Spermiogenic mRNAs are transcribed earlier and stored in a translationally inert state until needed for translation; however, it remains largely unclear how such repressed mRNAs become activated during spermiogenesis. We previously reported that the MIWI/piRNA machinery is responsible for mRNA elimination during late spermiogenesis in preparation for spermatozoa production. Here we unexpectedly discover that the same machinery is also responsible for activating translation of a subset of spermiogenic mRNAs to coordinate with morphological transformation into spermatozoa. Such action requires specific base-pairing interactions of piRNAs with target mRNAs in their 3' UTRs, which activates translation through coupling with cis-acting AU-rich elements to nucleate the formation of a MIWI/piRNA/eIF3f/HuR super-complex in a developmental stage-specific manner. These findings reveal a critical role of the piRNA system in translation activation, which we show is functionally required for spermatid development.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures


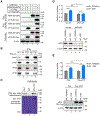



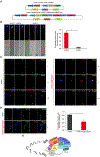
References
-
- Deng W, and Lin H (2002). miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819–830. - PubMed
-
- Gingras AC, Raught B, and Sonenberg N (1999). eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem 68, 913–963. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

