Deciphering miRNAs' Action through miRNA Editing
- PMID: 31835747
- PMCID: PMC6941098
- DOI: 10.3390/ijms20246249
Deciphering miRNAs' Action through miRNA Editing
Abstract
MicroRNAs (miRNAs) are small non-coding RNAs with the capability of modulating gene expression at the post-transcriptional level either by inhibiting messenger RNA (mRNA) translation or by promoting mRNA degradation. The outcome of a myriad of physiological processes and pathologies, including cancer, cardiovascular and metabolic diseases, relies highly on miRNAs. However, deciphering the precise roles of specific miRNAs in these pathophysiological contexts is challenging due to the high levels of complexity of their actions. Indeed, regulation of mRNA expression by miRNAs is frequently cell/organ specific; highly dependent on the stress and metabolic status of the organism; and often poorly correlated with miRNA expression levels. Such biological features of miRNAs suggest that various regulatory mechanisms control not only their expression, but also their activity and/or bioavailability. Several mechanisms have been described to modulate miRNA action, including genetic polymorphisms, methylation of miRNA promoters, asymmetric miRNA strand selection, interactions with RNA-binding proteins (RBPs) or other coding/non-coding RNAs. Moreover, nucleotide modifications (A-to-I or C-to-U) within the miRNA sequences at different stages of their maturation are also critical for their functionality. This regulatory mechanism called "RNA editing" involves specific enzymes of the adenosine/cytidine deaminase family, which trigger single nucleotide changes in primary miRNAs. These nucleotide modifications greatly influence a miRNA's stability, maturation and activity by changing its specificity towards target mRNAs. Understanding how editing events impact miRNA's ability to regulate stress responses in cells and organs, or the development of specific pathologies, e.g., metabolic diseases or cancer, should not only deepen our knowledge of molecular mechanisms underlying complex diseases, but can also facilitate the design of new therapeutic approaches based on miRNA targeting. Herein, we will discuss the current knowledge on miRNA editing and how this mechanism regulates miRNA biogenesis and activity.
Keywords: ADARs; APOBECs; miRNA editing; microRNA regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

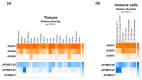
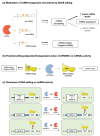
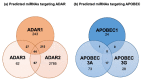
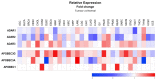
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

