Functional significance of U2AF1 S34F mutations in lung adenocarcinomas
- PMID: 31836708
- PMCID: PMC6911043
- DOI: 10.1038/s41467-019-13392-y
Functional significance of U2AF1 S34F mutations in lung adenocarcinomas
Abstract
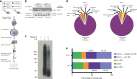
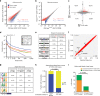
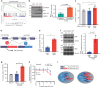
The functional role of U2AF1 mutations in lung adenocarcinomas (LUADs) remains incompletely understood. Here, we report a significant co-occurrence of U2AF1 S34F mutations with ROS1 translocations in LUADs. To characterize this interaction, we profiled effects of S34F on the transcriptome-wide distribution of RNA binding and alternative splicing in cells harboring the ROS1 translocation. Compared to its wild-type counterpart, U2AF1 S34F preferentially binds and modulates splicing of introns containing CAG trinucleotides at their 3' splice junctions. The presence of S34F caused a shift in cross-linking at 3' splice sites, which was significantly associated with alternative splicing of skipped exons. U2AF1 S34F induced expression of genes involved in the epithelial-mesenchymal transition (EMT) and increased tumor cell invasion. Finally, S34F increased splicing of the long over the short SLC34A2-ROS1 isoform, which was also associated with enhanced invasiveness. Taken together, our results suggest a mechanistic interaction between mutant U2AF1 and ROS1 in LUAD.
Conflict of interest statement
L.J.L. is an employee of and has ownership interests in 3 T Biosciences. N.I. is an employee of Genentech. A.M.N. is a co-inventor on patent applications related to ctDNA and other cancer biomarkers, has ownership interest in CiberMed, and has served as a consultant to CiberMed. S.V.B. is a co-inventor on patent applications related to cancer biomarkers. M.H.P. has ownership interests in and has served as a consultant for CRISPR Tx. H.Y.C. has ownership interests in Accent Therapeutics and has served as a consultant to 10X Genomics, Spring Discovery, and Accent Therapeutics. A.A.A. is a co-inventor on patent applications related to ctDNA and other cancer biomarkers, has ownership interests in CiberMed and FortySeven, and has served as a consultant/advisory board member for Genentech, Roche, Chugai, Gilead, Celgene, and CiberMed. M.D. is a co-inventor on patent applications related to ctDNA and other cancer biomarkers, has ownership interest in CiberMed, and has served as a consultant/advisory board member for Roche, AstraZeneca, Novartis, Quanticel, CiberMed, and BioNTech. All other authors have no conflicts of interest to declare.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

