Retrospective Validation of a Metagenomic Sequencing Protocol for Combined Detection of RNA and DNA Viruses Using Respiratory Samples from Pediatric Patients
- PMID: 31837435
- PMCID: PMC7106021
- DOI: 10.1016/j.jmoldx.2019.10.007
Retrospective Validation of a Metagenomic Sequencing Protocol for Combined Detection of RNA and DNA Viruses Using Respiratory Samples from Pediatric Patients
Abstract
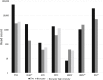
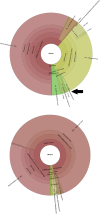
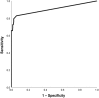
Viruses are the main cause of respiratory tract infections. Metagenomic next-generation sequencing (mNGS) enables unbiased detection of all potential pathogens. To apply mNGS in viral diagnostics, sensitive and simultaneous detection of RNA and DNA viruses is needed. Herein, were studied the performance of an in-house mNGS protocol for routine diagnostics of viral respiratory infections with potential for automated pan-pathogen detection. The sequencing protocol and bioinformatics analysis were designed and optimized, including exogenous internal controls. Subsequently, the protocol was retrospectively validated using 25 clinical respiratory samples. The developed protocol using Illumina NextSeq 500 sequencing showed high repeatability. Use of the National Center for Biotechnology Information's RefSeq database as opposed to the National Center for Biotechnology Information's nucleotide database led to enhanced specificity of classification of viral pathogens. A correlation was established between read counts and PCR cycle threshold value. Sensitivity of mNGS, compared with PCR, varied up to 83%, with specificity of 94%, dependent on the cutoff for defining positive mNGS results. Viral pathogens only detected by mNGS, not present in the routine diagnostic workflow, were influenza C, KI polyomavirus, cytomegalovirus, and enterovirus. Sensitivity and analytical specificity of this mNGS protocol were comparable to PCR and higher when considering off-PCR target viral pathogens. One single test detected all potential viral pathogens and simultaneously obtained detailed information on detected viruses.
Copyright © 2020 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Bates M., Mudenda V., Mwaba P., Zumla A. Deaths due to respiratory tract infections in Africa: a review of autopsy studies. Curr Opin Pulm Med. 2013;19:229–237. - PubMed
-
- Jain S., Self W.H., Wunderink R.G., Fakhran S., Balk R., Bramley A.M., Reed C., Grijalva C.G., Anderson E.J., Courtney D.M., Chappell J.D., Qi C., Hart E.M., Carroll F., Trabue C., Donnelly H.K., Williams D.J., Zhu Y., Arnold S.R., Ampofo K., Waterer G.W., Levine M., Lindstrom S., Winchell J.M., Katz J.M., Erdman D., Schneider E., Hicks L.A., McCullers J.A., Pavia A.T., Edwards K.M., Finelli L., CDC EPIC Study Team Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015;373:415–427. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

