DNA Facilitates Oligomerization and Prevents Aggregation via DNA Networks
- PMID: 31839258
- PMCID: PMC6952221
- DOI: 10.1016/j.bpj.2019.11.022
DNA Facilitates Oligomerization and Prevents Aggregation via DNA Networks
Abstract
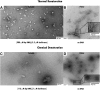
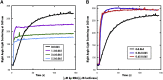
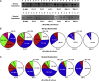
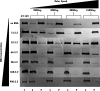
Previous studies have shown that nucleic acids can nucleate protein aggregation in disease-related proteins, but in other cases, they can act as molecular chaperones that prevent protein aggregation, even under extreme conditions. In this study, we describe the link between these two behaviors through a combination of electron microscopy and aggregation kinetics. We find that two different proteins become soluble under harsh conditions through oligomerization with DNA. These DNA/protein oligomers form "networks," which increase the speed of oligomerization. The cases of DNA both increasing and preventing protein aggregation are observed to stem from this enhanced oligomerization. This observation raises interesting questions about the role of nucleic acids in aggregate formation in disease states.
Copyright © 2019 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

