GCNA Interacts with Spartan and Topoisomerase II to Regulate Genome Stability
- PMID: 31839538
- PMCID: PMC7227305
- DOI: 10.1016/j.devcel.2019.11.006
GCNA Interacts with Spartan and Topoisomerase II to Regulate Genome Stability
Abstract
GCNA proteins are expressed across eukarya in pluripotent cells and have conserved functions in fertility. GCNA homologs Spartan (DVC-1) and Wss1 resolve DNA-protein crosslinks (DPCs), including Topoisomerase-DNA adducts, during DNA replication. Here, we show that GCNA mutants in mouse and C. elegans display defects in genome maintenance including DNA damage, aberrant chromosome condensation, and crossover defects in mouse spermatocytes and spontaneous genomic rearrangements in C. elegans. We show that GCNA and topoisomerase II (TOP2) physically interact in both mice and worms and colocalize on condensed chromosomes during mitosis in C. elegans embryos. Moreover, C. elegans gcna-1 mutants are hypersensitive to TOP2 poison. Together, our findings support a model in which GCNA provides genome maintenance functions in the germline and may do so, in part, by promoting the resolution of TOP2 DPCs.
Keywords: DNA-protein crosslink (DPC) repair; DVC-1; GCNA; Spartan; SprT; Top1; Top2; germ cells; topoisomerase.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures

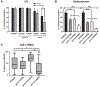
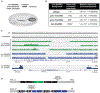
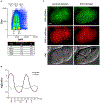

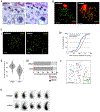

Comment in
-
When the Family Treasure Is a Doormat.Dev Cell. 2020 Jan 6;52(1):3-4. doi: 10.1016/j.devcel.2019.12.013. Dev Cell. 2020. PMID: 31951554
References
-
- Ahmed EA, Philippens ME, Kal HB, de Rooij DG, and de Boer P (2010). Genetic probing of homologous recombination and non-homologous end joining during meiotic prophase in irradiated mouse spermatocytes. Mutat. Res. 688, 12–18. - PubMed
-
- Ahmed S, and Hodgkin J (2000). MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403, 159–164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

