Ancient DNA suggests modern wolves trace their origin to a Late Pleistocene expansion from Beringia
- PMID: 31840921
- PMCID: PMC7317801
- DOI: 10.1111/mec.15329
Ancient DNA suggests modern wolves trace their origin to a Late Pleistocene expansion from Beringia
Abstract
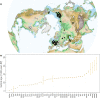
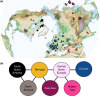
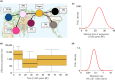
Grey wolves (Canis lupus) are one of the few large terrestrial carnivores that have maintained a wide geographical distribution across the Northern Hemisphere throughout the Pleistocene and Holocene. Recent genetic studies have suggested that, despite this continuous presence, major demographic changes occurred in wolf populations between the Late Pleistocene and early Holocene, and that extant wolves trace their ancestry to a single Late Pleistocene population. Both the geographical origin of this ancestral population and how it became widespread remain unknown. Here, we used a spatially and temporally explicit modelling framework to analyse a data set of 90 modern and 45 ancient mitochondrial wolf genomes from across the Northern Hemisphere, spanning the last 50,000 years. Our results suggest that contemporary wolf populations trace their ancestry to an expansion from Beringia at the end of the Last Glacial Maximum, and that this process was most likely driven by Late Pleistocene ecological fluctuations that occurred across the Northern Hemisphere. This study provides direct ancient genetic evidence that long-range migration has played an important role in the population history of a large carnivore, and provides insight into how wolves survived the wave of megafaunal extinctions at the end of the last glaciation. Moreover, because Late Pleistocene grey wolves were the likely source from which all modern dogs trace their origins, the demographic history described in this study has fundamental implications for understanding the geographical origin of the dog.
Keywords: Approximate Bayesian Computation; Pleistocene; ancient DNA; coalescent modelling; megafauna; population structure; population turnover; wolves.
© 2019 The Authors. Molecular Ecology published by John Wiley & Sons Ltd.
Figures


Comment in
-
Illuminating the mysteries of wolf history.Mol Ecol. 2020 May;29(9):1589-1591. doi: 10.1111/mec.15438. Epub 2020 May 19. Mol Ecol. 2020. PMID: 32286714
References
-
- Aggarwal, R. K. , Kivisild, T. , Ramadevi, J. , & Singh, L. (2007). Mitochondrial DNA coding region sequences support the phylogenetic distinction of two Indian wolf species. Journal of Zoological Systematics and Evolutionary Research, 45, 163–172. 10.1111/j.1439-0469.2006.00400.x - DOI
-
- Amante, C. , & Eakins, B. W. (2016). ETOPO1 1 Arc‐Minute Global Relief Model: Procedures, Data Sources and Analysis. NOAA Technical Memorandum NESDIS NGDC‐24. Retrieved from https://www.ngdc.noaa.gov/mgg/global/
-
- Baryshnikov, G. F. , Mol, D. , & Tikhonov, A. N. (2009). Finding of the Late Pleistocene carnivores in Taimyr Peninsula (Russia, Siberia) with paleoecological context. Russian Journal of Theriology, 8, 107–113. 10.15298/rusjtheriol.08.2.04 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

