Molecular epidemiology, genetic variability and evolution of HTLV-1 with special emphasis on African genotypes
- PMID: 31842895
- PMCID: PMC6916231
- DOI: 10.1186/s12977-019-0504-z
Molecular epidemiology, genetic variability and evolution of HTLV-1 with special emphasis on African genotypes
Abstract
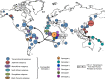
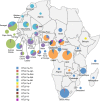
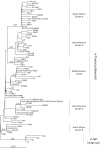
Human T cell leukemia virus (HTLV-1) is an oncoretrovirus that infects at least 10 million people worldwide. HTLV-1 exhibits a remarkable genetic stability, however, viral strains have been classified in several genotypes and subgroups, which often mirror the geographic origin of the viral strain. The Cosmopolitan genotype HTLV-1a, can be subdivided into geographically related subgroups, e.g. Transcontinental (a-TC), Japanese (a-Jpn), West-African (a-WA), North-African (a-NA), and Senegalese (a-Sen). Within each subgroup, the genetic diversity is low. Genotype HTLV-1b is found in Central Africa; it is the major genotype in Gabon, Cameroon and Democratic Republic of Congo. While strains from the HTLV-1d genotype represent only a few percent of the strains present in Central African countries, genotypes -e, -f, and -g have been only reported sporadically in particular in Cameroon Gabon, and Central African Republic. HTLV-1c genotype, which is found exclusively in Australo-Melanesia, is the most divergent genotype. This reflects an ancient speciation, with a long period of isolation of the infected populations in the different islands of this region (Australia, Papua New Guinea, Solomon Islands and Vanuatu archipelago). Until now, no viral genotype or subgroup is associated with a specific HTLV-1-associated disease. HTLV-1 originates from a simian reservoir (STLV-1); it derives from interspecies zoonotic transmission from non-human primates to humans (ancient or recent). In this review, we describe the genetic diversity of HTLV-1, and analyze the molecular mechanisms that are at play in HTLV-1 evolution. Similar to other retroviruses, HTLV-1 evolves either through accumulation of point mutations or recombination. Molecular studies point to a fairly low evolution rate of HTLV-1 (between 5.6E-7 and 1.5E-6 substitutions/site/year), supposedly because the virus persists within the host via clonal expansion (instead of new infectious cycles that use reverse transcriptase).
Keywords: Africa; Evolution; Genotypes; HTLV-1; Molecular epidemiology; Mutation rate.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Takatsuki K, Uchiyama T, Sagawa K, Yodoi J. Adult T cell leukemia in Japan. In: Seno S, Takaku F, Irino S, editors. Topics in hematology. Amsterdam: Excerpta Medica; 1977. pp. 73–77.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

