Acoustofluidic Salivary Exosome Isolation: A Liquid Biopsy Compatible Approach for Human Papillomavirus-Associated Oropharyngeal Cancer Detection
- PMID: 31843276
- PMCID: PMC6943372
- DOI: 10.1016/j.jmoldx.2019.08.004
Acoustofluidic Salivary Exosome Isolation: A Liquid Biopsy Compatible Approach for Human Papillomavirus-Associated Oropharyngeal Cancer Detection
Abstract
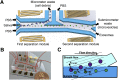
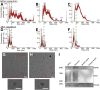
Previous efforts to evaluate the detection of human papilloma viral (HPV) DNA in whole saliva as a diagnostic measure for HPV-associated oropharyngeal cancer (HPV-OPC) have not shown sufficient clinical performance. We hypothesize that salivary exosomes are packaged with HPV-associated biomarkers, and efficient enrichment of salivary exosomes through isolation can enhance diagnostic and prognostic performance for HPV-OPC. In this study, an acoustofluidic (the fusion of acoustics and microfluidics) platform was developed to perform size-based isolation of salivary exosomes. These data showed that this platform is capable of consistently isolating exosomes from saliva samples, regardless of viscosity variation and collection method. Compared with the current gold standard, differential centrifugation, droplet digital RT-PCR analysis showed that the average yield of salivary exosomal small RNA from the acoustofluidic platform is 15 times higher. With this high-yield exosome isolation platform, we show that HPV16 DNA could be detected in isolated exosomes from the saliva of HPV-associated OPC patients at 80% concordance with tissues/biopsies positive for HPV16. Overall, these data demonstrated that the acoustofluidic platform can achieve high-purity and high-yield salivary exosome isolation for downstream salivary exosome-based liquid biopsy applications. Additionally, HPV16 DNA sequences in HPV-OPC patients are packaged in salivary exosomes and their isolation will enhance the detection of HPV16 DNA.
Copyright © 2020 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Wang M.B., Liu I.Y., Gornbein J.A., Nguyen C.T. HPV-positive oropharyngeal carcinoma: a systematic review of treatment and prognosis. Otolaryngol Head Neck Surg. 2015;153:758–769. - PubMed
-
- Sedghizadeh P.P., Billington W.D., Paxton D., Ebeed R., Mahabady S., Clark G.T., Enciso R. Is p16-positive oropharyngeal squamous cell carcinoma associated with favorable prognosis? A systematic review and meta-analysis. Oral Oncol. 2016;54:15–27. - PubMed
-
- Goy J., Hall S.F., Feldman-Stewart D., Groome P.A. Diagnostic delay and disease stage in head and neck cancer: a systematic review. Laryngoscope. 2009;119:889–898. - PubMed
-
- McGurk M., Scott S. The reality of identifying early oral cancer in the general dental practice. Br Dent J. 2010;208:347–351. - PubMed
-
- Zhan K.Y., Eskander A., Kang S.Y., Old M.O., Ozer E., Agrawal A.A., Carrau R.L., Rocco J.W., Teknos T.N. Appraisal of the AJCC 8th edition pathologic staging modifications for HPV− positive oropharyngeal cancer, a study of the National Cancer Data Base. Oral Oncol. 2017;73:152–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

