Divergent evolutionary trajectories of influenza B viruses underlie their contemporaneous epidemic activity
- PMID: 31843889
- PMCID: PMC6955377
- DOI: 10.1073/pnas.1916585116
Divergent evolutionary trajectories of influenza B viruses underlie their contemporaneous epidemic activity
Abstract
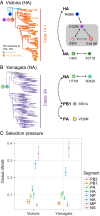
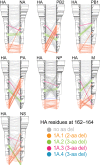
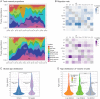
Influenza B viruses have circulated in humans for over 80 y, causing a significant disease burden. Two antigenically distinct lineages ("B/Victoria/2/87-like" and "B/Yamagata/16/88-like," termed Victoria and Yamagata) emerged in the 1970s and have cocirculated since 2001. Since 2015 both lineages have shown unusually high levels of epidemic activity, the reasons for which are unclear. By analyzing over 12,000 influenza B virus genomes, we describe the processes enabling the long-term success and recent resurgence of epidemics due to influenza B virus. We show that following prolonged diversification, both lineages underwent selective sweeps across the genome and have subsequently taken alternate evolutionary trajectories to exhibit epidemic dominance, with no reassortment between lineages. Hemagglutinin deletion variants emerged concomitantly in multiple Victoria virus clades and persisted through epistatic mutations and interclade reassortment-a phenomenon previously only observed in the 1970s when Victoria and Yamagata lineages emerged. For Yamagata viruses, antigenic drift of neuraminidase was a major driver of epidemic activity, indicating that neuraminidase-based vaccines and cross-reactivity assays should be employed to monitor and develop robust protection against influenza B morbidity and mortality. Overall, we show that long-term diversification and infrequent selective sweeps, coupled with the reemergence of hemagglutinin deletion variants and antigenic drift of neuraminidase, are factors that contributed to successful circulation of diverse influenza B clades. Further divergence of hemagglutinin variants with poor cross-reactivity could potentially lead to circulation of 3 or more distinct influenza B viruses, further complicating influenza vaccine formulation and highlighting the urgent need for universal influenza vaccines.
Keywords: antigenic; genetic diversity; natural selection; phylogeny; vaccine.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Shaw M. W., et al. , Reappearance and global spread of variants of influenza B/Victoria/2/87 lineage viruses in the 2000-2001 and 2001-2002 seasons. Virology 303, 1–8 (2002). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

