Multidrug Adaptive Resistance of Pseudomonas aeruginosa Swarming Cells
- PMID: 31844008
- PMCID: PMC7038238
- DOI: 10.1128/AAC.01999-19
Multidrug Adaptive Resistance of Pseudomonas aeruginosa Swarming Cells
Abstract
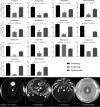
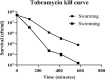
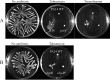
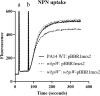
Swarming surface motility is a complex adaptation leading to multidrug antibiotic resistance and virulence factor production in Pseudomonas aeruginosa Here, we expanded previous studies to demonstrate that under swarming conditions, P. aeruginosa PA14 is more resistant to multiple antibiotics, including aminoglycosides, β-lactams, chloramphenicol, ciprofloxacin, tetracycline, trimethoprim, and macrolides, than swimming cells, but is not more resistant to polymyxin B. We investigated the mechanism(s) of swarming-mediated antibiotic resistance by examining the transcriptomes of swarming cells and swarming cells treated with tobramycin by transcriptomics (RNA-Seq) and reverse transcriptase quantitative PCR (qRT-PCR). RNA-Seq of swarming cells (versus swimming) revealed 1,581 dysregulated genes, including 104 transcriptional regulators, two-component systems, and sigma factors, numerous upregulated virulence and iron acquisition factors, and downregulated ribosomal genes. Strain PA14 mutants in resistome genes that were dysregulated under swarming conditions were tested for their ability to swarm in the presence of tobramycin. In total, 41 mutants in genes dysregulated under swarming conditions were shown to be more resistant to tobramycin under swarming conditions, indicating that swarming-mediated tobramycin resistance was multideterminant. Focusing on two genes downregulated under swarming conditions, both prtN and wbpW mutants were more resistant to tobramycin, while the prtN mutant was additionally resistant to trimethoprim under swarming conditions; complementation of these mutants restored susceptibility. RNA-Seq of swarming cells treated with subinhibitory concentrations of tobramycin revealed the upregulation of the multidrug efflux pump MexXY and downregulation of virulence factors.
Keywords: Pseudomonas aeruginosa; RNA-Seq; antibiotic resistance; swarming motility; tobramycin.
Copyright © 2020 American Society for Microbiology.
Figures


References
-
- Cystic Fibrosis Foundation. 2016. 2016 Patient registry annual data report. Cystic Fibrosis Foundation, Bethesda, MD.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

