Artificial signaling in mammalian cells enabled by prokaryotic two-component system
- PMID: 31844302
- PMCID: PMC6982536
- DOI: 10.1038/s41589-019-0429-9
Artificial signaling in mammalian cells enabled by prokaryotic two-component system
Abstract
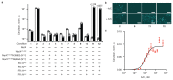
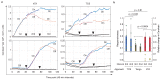
Augmenting live cells with new signal transduction capabilities is a key objective in genetic engineering and synthetic biology. We showed earlier that two-component signaling pathways could function in mammalian cells, albeit while losing their ligand sensitivity. Here, we show how to transduce small-molecule ligands in a dose-dependent fashion into gene expression in mammalian cells using two-component signaling machinery. First, we engineer mutually complementing truncated mutants of a histidine kinase unable to dimerize and phosphorylate the response regulator. Next, we fuse these mutants to protein domains capable of ligand-induced dimerization, which restores the phosphoryl transfer in a ligand-dependent manner. Cytoplasmic ligands are transduced by facilitating mutant dimerization in the cytoplasm, while extracellular ligands trigger dimerization at the inner side of a plasma membrane. These findings point to the potential of two-component regulatory systems as enabling tools for orthogonal signaling pathways in mammalian cells.
Conflict of interest statement
A patent application has been filed describing the result in this study, with A.M. and Y.B. listed as coinventors. Y.B. is a coinventor of a background patent to this filing.
Figures




Comment in
-
Mammalian signaling circuits from bacterial parts.Nat Chem Biol. 2020 Feb;16(2):110-111. doi: 10.1038/s41589-019-0436-x. Nat Chem Biol. 2020. PMID: 31844303 No abstract available.
References
-
- Park SH, Zarrinpar A, Lim WA. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. - PubMed
-
- Benenson Y. Biomolecular computing systems: principles, progress and potential. Nat Rev Genet. 2012;13:455–468. - PubMed
-
- Hansen J, Benenson Y. Synthetic biology of cell signaling. Nat Comput. 2016;15:5–13.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

