High-throughput phenotyping reveals expansive genetic and structural underpinnings of immune variation
- PMID: 31844327
- PMCID: PMC7338221
- DOI: 10.1038/s41590-019-0549-0
High-throughput phenotyping reveals expansive genetic and structural underpinnings of immune variation
Abstract
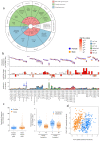
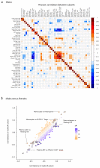
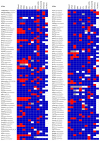
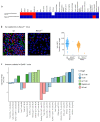
By developing a high-density murine immunophenotyping platform compatible with high-throughput genetic screening, we have established profound contributions of genetics and structure to immune variation (http://www.immunophenotype.org). Specifically, high-throughput phenotyping of 530 unique mouse gene knockouts identified 140 monogenic 'hits', of which most had no previous immunologic association. Furthermore, hits were collectively enriched in genes for which humans show poor tolerance to loss of function. The immunophenotyping platform also exposed dense correlation networks linking immune parameters with each other and with specific physiologic traits. Such linkages limit freedom of movement for individual immune parameters, thereby imposing genetically regulated 'immunologic structures', the integrity of which was associated with immunocompetence. Hence, we provide an expanded genetic resource and structural perspective for understanding and monitoring immune variation in health and disease.
Figures



References
Publication types
MeSH terms
Grants and funding
- FC001093/WT_/Wellcome Trust/United Kingdom
- R01 GM118417/GM/NIGMS NIH HHS/United States
- MC_UU_00008/6/MRC_/Medical Research Council/United Kingdom
- FC001093/CRUK_/Cancer Research UK/United Kingdom
- 102972/WT_/Wellcome Trust/United Kingdom
- R37 AI026170/AI/NIAID NIH HHS/United States
- MR/R007748/1/MRC_/Medical Research Council/United Kingdom
- 20265/CRUK_/Cancer Research UK/United Kingdom
- 107059/WT_/Wellcome Trust/United Kingdom
- 210661/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- R01 AI026170/AI/NIAID NIH HHS/United States
- UM1 HG006370/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

