77Se NMR Probes the Protein Environment of Selenomethionine
- PMID: 31846581
- PMCID: PMC8088340
- DOI: 10.1021/acs.jpcb.9b07466
77Se NMR Probes the Protein Environment of Selenomethionine
Abstract
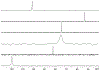
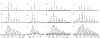
Sulfur is critical for the correct structure and proper function of proteins. Yet, lacking a sensitive enough isotope, nuclear magnetic resonance (NMR) experiments are unable to deliver for sulfur in proteins the usual wealth of chemical, dynamic, and structural information. This limitation can be circumvented by substituting sulfur with selenium, which has similar physicochemical properties and minimal impact on protein structures but possesses an NMR compatible isotope (77Se). Here we exploit the sensitivity of 77Se NMR to the nucleus' chemical milieu and use selenomethionine as a probe for its proteinaceous environment. However, such selenium NMR spectra of proteins currently resist a reliable interpretation because systematic connections between variations of system variables and changes in 77Se NMR parameters are still lacking. To start narrowing this knowledge gap, we report here on a biological 77Se magnetic resonance data bank based on a systematically designed library of GB1 variants in which a single selenomethionine was introduced at different locations within the protein. We recorded the resulting isotropic 77Se chemical shifts and relaxation times for six GB1 variants by solution-state 77Se NMR. For four of the GB1 variants we were also able to determine the chemical shift anisotropy tensor of SeM by solid-state 77Se NMR. To enable interpretation of the NMR data, the structures of five of the GB1 variants were solved by X-ray crystallography to a resolution of 1.2 Å, allowing us to unambiguously determine the conformation of the selenomethionine. Finally, we combine our solution- and solid-state NMR data with the structural information to arrive at general insights regarding the execution and interpretation of 77Se NMR experiments that exploit selenomethionine to probe proteins.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
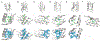
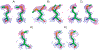



References
-
- Beno BR; Yeung KS; Bartberger MD; Pennington LD; Meanwell NA A survey of the role of noncovalent sulfur interactions in drug design. J. Med. Chem 2015, 58 (11), 4383–438. - PubMed
-
- O’Dell LA; Moudrakovski IL Testing the sensitivity limits of S-33 NMR: An ultra-wideline study of elemental sulfur. J. Magn. Reson 2010, 207 (2), 345–347. - PubMed
-
- Wessjohann LA; Schneider A; Abbas M; Brandt W Selenium in chemistry and biochemistry in comparison to sulfur. Biol. Chem 2007, 388 (10), 997–1006. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

