Self-Organization of Mouse Stem Cells into an Extended Potential Blastoid
- PMID: 31846649
- PMCID: PMC10291877
- DOI: 10.1016/j.devcel.2019.11.014
Self-Organization of Mouse Stem Cells into an Extended Potential Blastoid
Abstract
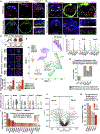
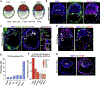
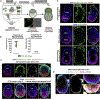
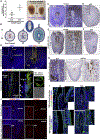
Mammalian blastocysts comprise three distinct cell lineages essential for development beyond implantation: the pluripotent epiblast, which generates the future embryo, and surrounding it the extra-embryonic primitive endoderm and the trophectoderm tissues. Embryonic stem cells can reintegrate into embryogenesis but contribute primarily to epiblast lineages. Here, we show that mouse embryonic stem cells cultured under extended pluripotent conditions (EPSCs) can be partnered with trophoblast stem cells to self-organize into blastocyst-like structures with all three embryonic and extra-embryonic lineages. Morphogenetic and transcriptome profiling analyses reveal that these blastocyst-like structures show distinct embryonic-abembryonic axes and primitive endoderm differentiation and can initiate the transition from the pre- to post-implantation egg cylinder morphology in vitro.
Keywords: Blastoid; Development; Embryo; Endoderm; Extended pluripotency; In Vitro; Preimplantation; Stem cells; Trophoblast.
Published by Elsevier Inc.
Figures



References
-
- Anderson KGV, Hamilton WB, Roske FV, Azad A, Knudsen TE, Canham MA, Forrester LM, and Brickman JM (2017). Insulin fine-tunes self-renewal pathways governing naive pluripotency and extra-embryonic endoderm. Nat. Cell Biol 19, 1164–1177. - PubMed
-
- Andrews S (2010). FastQC a quality control to tool for high throughput sequence data
-
- Aplin JD, and Ruane PT (2017). Embryo-epithelium interactions during implantation at a glance. J. Cell Sci 130, 15–22. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

