Detection of ALDH3B2 in Human Placenta
- PMID: 31847104
- PMCID: PMC6941052
- DOI: 10.3390/ijms20246292
Detection of ALDH3B2 in Human Placenta
Abstract
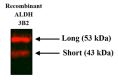
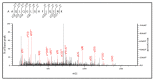
Aldehyde dehydrogenase 3B2 (ALDH3B2) gene contains a premature termination codon, which can be skipped or suppressed resulting in full-length protein expression. Alternatively, the longest putative open reading frame starting with the second in-frame start codon would encode short isoform. No unequivocal evidence of ALDH3B2 expression in healthy human tissues is available. The aim of this study was to confirm its expression in human placenta characterized by the highest ALDH3B2 mRNA abundance. ALDH3B2 DNA and mRNA were sequenced. The expression was investigated using western blot. The identity of the protein was confirmed using mass spectrometry (MS). The predicted tertiary and quaternary structures, subcellular localization, and phosphorylation sites were assessed using bioinformatic analyses. All DNA and mRNA isolates contained the premature stop codon. In western blot analyses, bands corresponding to the mass of full-length protein were detected. MS analysis led to the identification of two unique peptides, one of which is encoded by the nucleotide sequence located upstream the second start codon. Bioinformatic analyses suggest cytoplasmic localization and several phosphorylation sites. Despite premature stop codon in DNA and mRNA sequences, full-length ALDH3B2 was found. It can be formed as a result of premature stop codon readthrough, complex phenomenon enabling stop codon circumvention.
Keywords: ALDH3B2; placenta; premature stop codon; readthrough.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








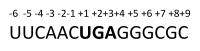
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

