The Peptide Venom Composition of the Fierce Stinging Ant Tetraponera aethiops (Formicidae: Pseudomyrmecinae)
- PMID: 31847368
- PMCID: PMC6950161
- DOI: 10.3390/toxins11120732
The Peptide Venom Composition of the Fierce Stinging Ant Tetraponera aethiops (Formicidae: Pseudomyrmecinae)
Abstract
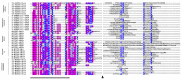
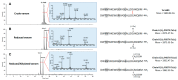
In the mutualisms involving certain pseudomyrmicine ants and different myrmecophytes (i.e., plants sheltering colonies of specialized "plant-ant" species in hollow structures), the ant venom contributes to the host plant biotic defenses by inducing the rapid paralysis of defoliating insects and causing intense pain to browsing mammals. Using integrated transcriptomic and proteomic approaches, we identified the venom peptidome of the plant-ant Tetraponera aethiops (Pseudomyrmecinae). The transcriptomic analysis of its venom glands revealed that 40% of the expressed contigs encoded only seven peptide precursors related to the ant venom peptides from the A-superfamily. Among the 12 peptide masses detected by liquid chromatography-mass spectrometry (LC-MS), nine mature peptide sequences were characterized and confirmed through proteomic analysis. These venom peptides, called pseudomyrmecitoxins (PSDTX), share amino acid sequence identities with myrmeciitoxins known for their dual offensive and defensive functions on both insects and mammals. Furthermore, we demonstrated through reduction/alkylation of the crude venom that four PSDTXs were homo- and heterodimeric. Thus, we provide the first insights into the defensive venom composition of the ant genus Tetraponera indicative of a streamlined peptidome.
Keywords: Tetraponera aethiops; defensive venom; dimeric peptides; peptidome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Venom toxicity and composition in three Pseudomyrmex ant species having different nesting modes.Toxicon. 2014 Sep;88:67-76. doi: 10.1016/j.toxicon.2014.05.022. Epub 2014 Jun 11. Toxicon. 2014. PMID: 24929139
-
Comparisons of Protein and Peptide Complexity in Poneroid and Formicoid Ant Venoms.J Proteome Res. 2016 Sep 2;15(9):3039-54. doi: 10.1021/acs.jproteome.6b00182. Epub 2016 Aug 2. J Proteome Res. 2016. PMID: 27436154
-
The complexity and structural diversity of ant venom peptidomes is revealed by mass spectrometry profiling.Rapid Commun Mass Spectrom. 2015 Mar 15;29(5):385-96. doi: 10.1002/rcm.7116. Rapid Commun Mass Spectrom. 2015. PMID: 26349460
-
Diversity of peptide toxins from stinging ant venoms.Toxicon. 2014 Dec 15;92:166-78. doi: 10.1016/j.toxicon.2014.10.021. Epub 2014 Oct 28. Toxicon. 2014. PMID: 25448389 Review.
-
The Biochemical Toxin Arsenal from Ant Venoms.Toxins (Basel). 2016 Jan 20;8(1):30. doi: 10.3390/toxins8010030. Toxins (Basel). 2016. PMID: 26805882 Free PMC article. Review.
Cited by
-
Studying the Rationale of Fire Ant Sting Therapy Usage by the Tribal Natives of Bastar Revealed Ant Venom-Derived Peptides with Promising Anti-Malarial Activity.Toxins (Basel). 2022 Nov 11;14(11):789. doi: 10.3390/toxins14110789. Toxins (Basel). 2022. PMID: 36422964 Free PMC article.
-
Venomics of the Central European Myrmicine Ants Myrmica rubra and Myrmica ruginodis.Toxins (Basel). 2022 May 21;14(5):358. doi: 10.3390/toxins14050358. Toxins (Basel). 2022. PMID: 35622604 Free PMC article.
-
Nature-inspired dimerization as a strategy to modulate neuropeptide pharmacology exemplified with vasopressin and oxytocin.Chem Sci. 2021 Feb 4;12(11):4057-4062. doi: 10.1039/d0sc05501h. Chem Sci. 2021. PMID: 34163676 Free PMC article.
-
Anti-inflammatory activities of arthropod peptides: a systematic review.J Venom Anim Toxins Incl Trop Dis. 2021 Oct 22;27:e20200152. doi: 10.1590/1678-9199-JVATITD-2020-0152. eCollection 2021. J Venom Anim Toxins Incl Trop Dis. 2021. PMID: 34795699 Free PMC article. Review.
-
Composition and Acute Inflammatory Response from Tetraponera rufonigra Venom on RAW 264.7 Macrophage Cells.Toxins (Basel). 2021 Apr 3;13(4):257. doi: 10.3390/toxins13040257. Toxins (Basel). 2021. PMID: 33916734 Free PMC article.
References
-
- Blank S., Seismann H., Bockisch B., Braren I., Cifuentes L., McIntyre M., Ruhl D., Ring J., Bredehorst R., Ollert M.W., et al. Identification, recombinant expression, and characterization of the 100 kDa high molecular weight hymenoptera venom allergens Api m 5 and Ves v 3. J. Immunol. 2010;184:5403–5413. doi: 10.4049/jimmunol.0803709. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

