Metabolomics: A Way Forward for Crop Improvement
- PMID: 31847393
- PMCID: PMC6969922
- DOI: 10.3390/metabo9120303
Metabolomics: A Way Forward for Crop Improvement
Abstract
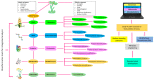
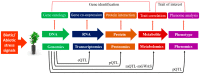
Metabolomics is an emerging branch of "omics" and it involves identification and quantification of metabolites and chemical footprints of cellular regulatory processes in different biological species. The metabolome is the total metabolite pool in an organism, which can be measured to characterize genetic or environmental variations. Metabolomics plays a significant role in exploring environment-gene interactions, mutant characterization, phenotyping, identification of biomarkers, and drug discovery. Metabolomics is a promising approach to decipher various metabolic networks that are linked with biotic and abiotic stress tolerance in plants. In this context, metabolomics-assisted breeding enables efficient screening for yield and stress tolerance of crops at the metabolic level. Advanced metabolomics analytical tools, like non-destructive nuclear magnetic resonance spectroscopy (NMR), liquid chromatography mass-spectroscopy (LC-MS), gas chromatography-mass spectrometry (GC-MS), high performance liquid chromatography (HPLC), and direct flow injection (DFI) mass spectrometry, have sped up metabolic profiling. Presently, integrating metabolomics with post-genomics tools has enabled efficient dissection of genetic and phenotypic association in crop plants. This review provides insight into the state-of-the-art plant metabolomics tools for crop improvement. Here, we describe the workflow of plant metabolomics research focusing on the elucidation of biotic and abiotic stress tolerance mechanisms in plants. Furthermore, the potential of metabolomics-assisted breeding for crop improvement and its future applications in speed breeding are also discussed. Mention has also been made of possible bottlenecks and future prospects of plant metabolomics.
Keywords: abiotic stress; biotic stress; crop improvement; mass spectrometry; metabolic profiling; metabolomics; metabolomics-assisted breeding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Obata T., Witt S., Lisec J., Palacios-Rojas N., Florez-Sarasa I., Yousfi S., Araus J.L., Cairns J.E., Fernie A.R. Metabolite profiles of maize leaves in drought, heat, and combined stress field trials reveal the relationship between metabolism and grain yield. Plant Physiol. 2015;169:2665–2683. doi: 10.1104/pp.15.01164. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

