Long non-coding RNA BZRAP1-AS1 silencing suppresses tumor angiogenesis in hepatocellular carcinoma by mediating THBS1 methylation
- PMID: 31847842
- PMCID: PMC6916030
- DOI: 10.1186/s12967-019-02145-6
Long non-coding RNA BZRAP1-AS1 silencing suppresses tumor angiogenesis in hepatocellular carcinoma by mediating THBS1 methylation
Abstract
Background: Hepatocellular carcinoma (HCC) is the most frequent primary liver cancer associated with a high mortality. Long non-coding RNAs (lncRNAs) have recently emerged as regulators in the development and progression of several cancers, and therefore represent an opportunity to uncover new targets for therapy. In the present study, we aimed to investigate the potential effect of lncRNA BZRAP1-AS1 on the angiogenesis of HCC.
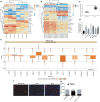
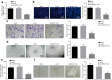
Methods: Microarray-based data analysis was initially employed to screen genes and lncRNAs that are differentially expressed in HCC and the candidate BZRAP1-AS1 was identified as a hit. The expression of BZRAP1-AS1 and thrombospondin-1 (THBS1) in HCC tissues and cells were then determined using RT-qPCR. The gene methylation level was measured by methylation-specific PCR (MSP) and bisulfite sequencing PCR (BSP) assays. Next, the interactions between BZRAP1-AS1, DNA methyltransferase 3B (DNMT3b), and THBS1 were assessed by RIP, RNA pull-down and ChIP assays. Finally, the roles of BZRAP1-AS1, DNMT3b and THBS1 in angiogenesis in vitro as well as tumorigenesis in vivo were evaluated by a battery of the gain- and loss-of function experiments.
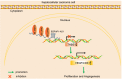
Results: BZRAP1-AS1 was identified as a highly expressed lncRNA in HCC tissues and cells. Down-regulation of BZRAP1-AS1 in HCC cells inhibited HUVEC proliferation, migration and angiogenesis. By interacting with DNMT3b, BZRAP1-AS1 induced methylation of the THBS1 promoter and inhibited the transcription of THBS1, resulting in promoted angiogenesis of HUVECs. Moreover, silencing of BZRAP1-AS1 repressed the angiogenesis as well as the tumor growth of HCC in vivo via up-regulating THBS1.
Conclusion: This study provides evidence that angiogenesis in HCC is hindered by silencing of BZRAP1-AS1. Thus, BZRAP1-AS1 may be a promising marker for the treatment of HCC.
Keywords: Angiogenesis; BZRAP1-AS1; DNA methylation; DNA methyltransferase 3B; Hepatocellular carcinoma; Long non-coding RNA; Thrombospondin-1.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

