Retroconversion of estrogens into androgens by bacteria via a cobalamin-mediated methylation
- PMID: 31848239
- PMCID: PMC6983444
- DOI: 10.1073/pnas.1914380117
Retroconversion of estrogens into androgens by bacteria via a cobalamin-mediated methylation
Abstract
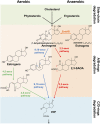
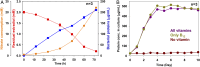
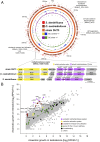
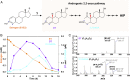
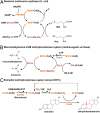
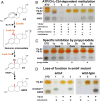
Steroid estrogens modulate physiology and development of vertebrates. Conversion of C19 androgens into C18 estrogens is thought to be an irreversible reaction. Here, we report a denitrifying Denitratisoma sp. strain DHT3 capable of catabolizing estrogens or androgens anaerobically. Strain DHT3 genome contains a polycistronic gene cluster, emtABCD, differentially transcribed under estrogen-fed conditions and predicted to encode a cobalamin-dependent methyltransferase system conserved among estrogen-utilizing anaerobes; an emtA-disrupted DHT3 derivative could catabolize androgens but not estrogens. These data, along with the observed androgen production in estrogen-fed strain DHT3 cultures, suggested the occurrence of a cobalamin-dependent estrogen methylation to form androgens. Consistently, the estrogen conversion into androgens in strain DHT3 cell extracts requires methylcobalamin and is inhibited by propyl iodide, a specific inhibitor of cobalamin-dependent enzymes. The identification of the cobalamin-dependent estrogen methylation thus represents an unprecedented metabolic link between cobalamin and steroid metabolism and suggests that retroconversion of estrogens into androgens occurs in the biosphere.
Keywords: biocatalysis; cobalamin-dependent methyltransferase; estrogens; microbial metabolism; steroids.
Conflict of interest statement
The authors declare no competing interest.
Figures

Comment in
-
Anaerobic bacteria need their vitamin B12 to digest estrogen.Proc Natl Acad Sci U S A. 2020 Jan 28;117(4):1833-1835. doi: 10.1073/pnas.1921340117. Epub 2020 Jan 9. Proc Natl Acad Sci U S A. 2020. PMID: 31919281 Free PMC article. No abstract available.
References
-
- Albrecht E. D., Pepe G. J., Steroid hormone regulation of angiogenesis in the primate endometrium. Front. Biosci. 8, d416–d429 (2003). - PubMed
-
- Ryan K. J., Biochemistry of aromatase: Significance to female reproductive physiology. Cancer Res. 42 (suppl. 8), 3342s–3344s (1982). - PubMed
-
- Mechoulam R., Brueggemeier R., Denlinger D., Estrogens in insects. Cell. Mol. Life Sci. 40, 942–944 (1984).
-
- Tarrant A. M., Blomquist C. H., Lima P. H., Atkinson M. J., Atkinson S., Metabolism of estrogens and androgens by scleractinian corals. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 136, 473–485 (2003). - PubMed
-
- Griffith D. R., Kido Soule M. C., Eglinton T. I., Kujawinski E. B., Gschwend P. M., Steroidal estrogen sources in a sewage-impacted coastal ocean. Environ. Sci. Process. Impacts 18, 981–991 (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

