Diversity in Natural Transformation Frequencies and Regulation across Vibrio Species
- PMID: 31848285
- PMCID: PMC6918086
- DOI: 10.1128/mBio.02788-19
Diversity in Natural Transformation Frequencies and Regulation across Vibrio Species
Abstract
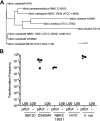
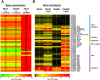
In Vibrio species, chitin-induced natural transformation enables bacteria to take up DNA from the external environment and integrate it into their genome. Expression of the master competence regulator TfoX bypasses the need for chitin induction and drives expression of the genes required for competence in several Vibrio species. Here, we show that TfoX expression in Vibrio campbellii strains DS40M4 and NBRC 15631 enables high natural transformation frequencies. Conversely, transformation was not achieved in the model quorum-sensing strain V. campbellii BB120 (previously classified as Vibrio harveyi). Surprisingly, we find that quorum sensing is not required for transformation in V. campbellii DS40M4 or Vibrio parahaemolyticus in contrast to the established regulatory pathway in Vibrio cholerae in which quorum sensing is required to activate the competence regulator QstR. Similar to V. cholerae, expression of both QstR and TfoX is necessary for transformation in DS40M4. There is a wide disparity in transformation frequencies among even closely related Vibrio strains, with V. vulnificus having the lowest functional transformation frequency. Ectopic expression of both TfoX and QstR is sufficient to produce a significant increase in transformation frequency in Vibrio vulnificus To explore differences in competence regulation, we used previously studied V. cholerae competence genes to inform a comparative genomics analysis coupled with transcriptomics. We find that transformation capability cannot necessarily be predicted by the level of gene conservation but rather correlates with competence gene expression following TfoX induction. Thus, we have uncovered notable species- and strain-level variations in the competence gene regulation pathway across the Vibrio genus.IMPORTANCE Naturally transformable, or competent, bacteria are able to take up DNA from their environment, a key method of horizontal gene transfer for acquisition of new DNA sequences. Our research shows that Vibrio species that inhabit marine environments exhibit a wide diversity in natural transformation capability ranging from nontransformability to high transformation rates in which 10% of cells measurably incorporate new DNA. We show that the role of regulatory systems controlling the expression of competence genes (e.g., quorum sensing) differs throughout both the species and strain levels. We explore natural transformation capabilities of Vibrio campbellii species which have been thus far uncharacterized and find novel regulation of competence. Expression of two key transcription factors, TfoX and QstR, is necessary to stimulate high levels of transformation in Vibrio campbellii and recover low rates of transformation in Vibrio vulnificus.
Keywords: competence; natural transformation; quorum sensing; vibrio.
Copyright © 2019 Simpson et al.
Figures





References
-
- Yamamoto S, Izumiya H, Mitobe J, Morita M, Arakawa E, Ohnishi M, Watanabe H. 2011. Identification of a chitin-induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae. J Bacteriol 193:1953–1965. doi:10.1128/JB.01340-10. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

