Characterization of the Pig Gut Microbiome and Antibiotic Resistome in Industrialized Feedlots in China
- PMID: 31848308
- PMCID: PMC6918024
- DOI: 10.1128/mSystems.00206-19
Characterization of the Pig Gut Microbiome and Antibiotic Resistome in Industrialized Feedlots in China
Abstract
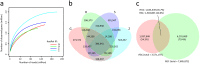
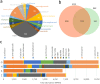
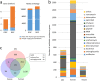
To characterize the diversity and richness and explore the function and structure of swine gut microbiome and resistome in common pig-farming feedlots, we sampled and metagenomic sequenced the feces of pigs from four different industrialized feedlots located in four distant provinces across China. Surprisingly, more than half of the nonredundant genes (1,937,648, 54.3%) in the current catalogue were newly found compared with the previously published reference gene catalogue (RGC) of the pig gut microbiome. Additionally, 16 high-completeness draft genomes were obtained by analyzing the dominant species on each feedlot. Notably, seven of these species often appeared in the human body sites. Despite a smaller number of nonredundant genes, our study identified more antibiotic resistance genes than those available in the RGC. Tetracycline, aminoglycoside, and multidrug resistance genes accounted for nearly 70% of the relative abundance in the current catalogue. Slightly higher sharing ratios were shown between the industrialized feedlot pig gut microbiomes and human gut microbiomes than that between the RGC and human counterpart (14.7% versus 12.6% in genes and 94.1% versus 87.7% in functional groups, respectively). Furthermore, a remarkably high number of the antibiotic resistance proteins (n =141) were identified to be shared by the pig, human, and mouse resistome, indicating the potential for horizontal transfer of resistance genes. Of the antibiotic resistance proteins shared by pigs and humans, 50 proteins were related to tetracycline resistance, and 49 were related to aminoglycoside resistance.IMPORTANCE The gut microbiota is believed to be closely related to many important physical functions in the host. Comprehensive data on mammalian gut metagenomes has facilitated research on host-microbiome interaction mechanisms, but less is known about pig gut microbiome, especially the gut microbiome in industrialized feedlot pigs, compared with human microbiome. On the other hand, pig production, as an important source of food, is believed to exacerbate the antibiotic resistance in humans due to the abuse of antibiotics in pig production in various parts of the world. This study delineates an intricate picture of swine gut microbiome and antibiotic resistome in industrialized feedlots and may provide insight for the pig producing industry.
Keywords: antibiotic resistance gene; antibiotic resistome; gene catalogue; industrialized feedlot; pig gut microbiome.
Copyright © 2019 Wang et al.
Figures

References
LinkOut - more resources
Full Text Sources

